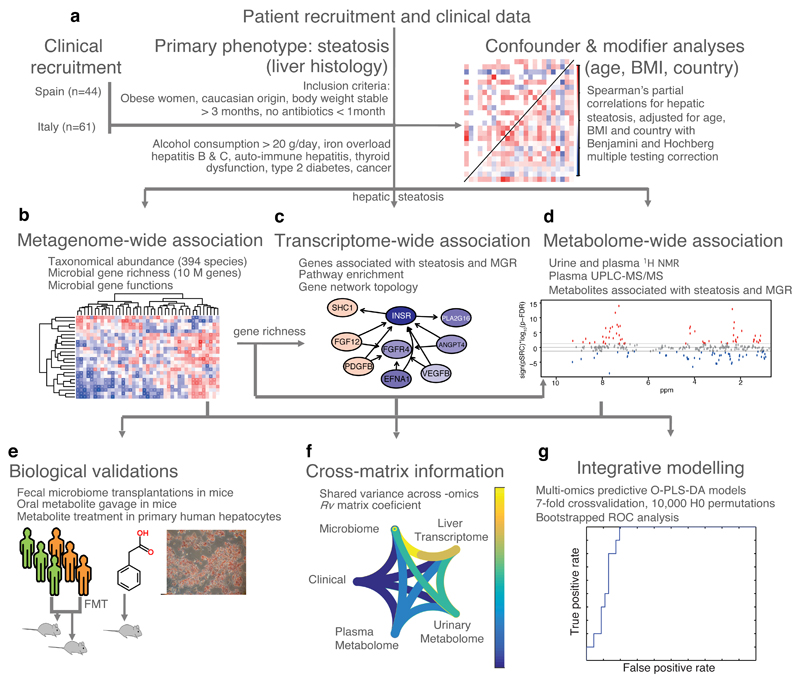
Figure 1. Flowchart showing approach used for the integration of clinical, molecular phenomics and metagenomic information and biological validations.
a, Confounder and modifier analysis performed using linear models on the FLORINASH clinical markers identified three confounders: age, BMI and country (n = 105). Subsequent analyses were performed using partial Spearman’s rank-based correlation (pSRC) coefficients adjusted for age, BMI and country and corrected for multiple testing using the Benjamini and Hochberg criterion (p-FDR). b, Metagenome-wide and phenome-wide association of taxonomic abundance data with clinical markers (n = 56 patients, pSRC, p-FDR < 0.05). c, Network analysis of hepatic transcriptome (n = 56 patients, pSRC, p-FDR < 0.05). d, Metabolome-Wide Association Study based on plasma (n = 56) and urine (n = 102, pSRC, p-FDR < 0.05) 1H-NMR spectra. e, In vitro and in vivo pre-clinical validation protocols. f, Integrative comparison analysis using Rv coefficients (n = 56). g, Predictive performance of an O-PLS-DA model integrating all metagenomic and phenomic modalities for prediction of non-alcoholic fatty liver (no hepatic steatosis, score = 0, n = 10 vs. steatosis, score > 0, n = 46) in ROC curves. All tests are two-sided.