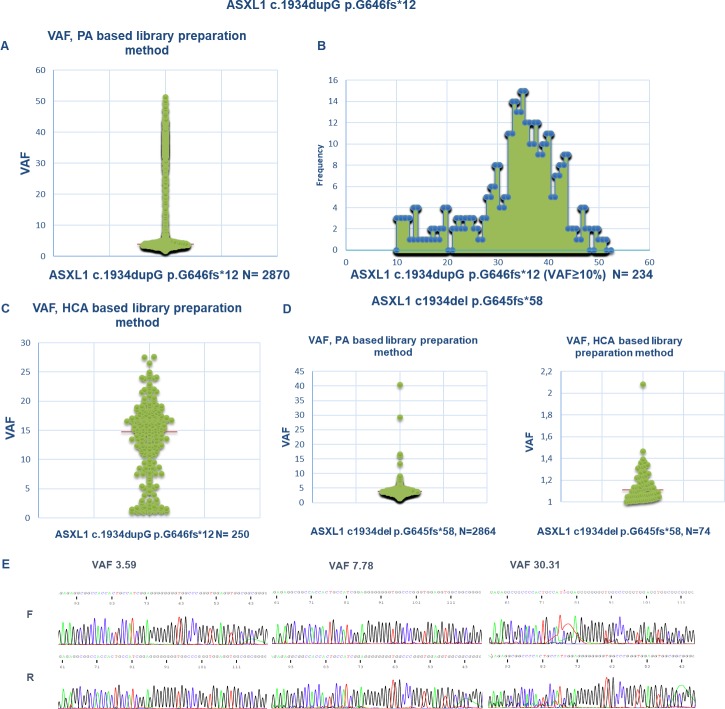
Fig 1.
A. VAF scatterplot plotting all the clinical samples studied by NGS with direct PCR amplification (PA) based library preparation chemistry found to harbor ASXL1 c.1934dupG (n = 2870). Majority of the cases (2636, 92%) have a variant frequency below 10%. Y axis: VAF. B. 234 samples (8%) were identified with ASXL1 c.1934dupG with VAF≥10%. The maximum VAF observed in the series was 51.4. X axis: VAF, Y axis: number of samples. C. VAF scatterplot showing all the clinical samples studied by HCA-based NGS library preparation chemistry found to harbor ASXL1 c.1934dupG (n = 250).Y axis: VAF D. Left panel: VAF scatterplot showing all the clinical samples studied by NGS with amplicon based library preparation chemistry found to harbor ASXL1 c.1934del p.G645fs (n = 2864). As depicted, most of the cases (99%) have a variant frequency below 10%. The maximum VAF observed in the series was 40.36. Y axis: VAF. Right panel: VAF scatterplot showing all the clinical samples studied by HCA-based NGS found to harbor ASXL1 c.1934del p.G645fs (n = 74). Y axis: VAF. E. Three electropherograms of cases with ASXL1 c.1934dupG quantified as 3.59, 7.78 and 30.31 respectively by NGS after amplicon based library preparation. As shown, no minor clonal sequence is found in the case with a VAF<5%.