Abstract
Idiopathic pulmonary fibrosis (IPF) is a devastating fibrotic lung disease of unknown etiology and limited therapeutic options. In this report, we characterize what we believe is a novel CCR10+ epithelial cell population in IPF lungs. There was a significant increase in the percentage of CCR10+ epithelial cells in IPF relative to normal lung explants and their numbers significantly correlated to lung remodeling in humanized NSG mice. Cultured CCR10-enriched IPF epithelial cells promoted IPF lung fibroblast invasion and collagen 1 secretion. Single-cell RNA sequencing analysis showed distinct CCR10+ epithelial cell populations enriched for inflammatory and profibrotic transcripts. Consistently, cultured IPF but not normal epithelial cells induced lung remodeling in humanized NSG mice, where the number of CCR10+ IPF, but not normal, epithelial cells correlated with hydroxyproline concentration in the remodeled NSG lungs. A subset of IPF CCR10hi epithelial cells coexpress EphA3 and ephrin A signaling induces the expression of CCR10 by these cells. Finally, EphA3+CCR10hi epithelial cells induce more consistent lung remodeling in NSG mice relative to EphA3–CCR10lo epithelial cells. Our results suggest that targeting epithelial cells, highly expressing CCR10, may be beneficial in IPF.
Keywords: Cell Biology, Pulmonology
Keywords: Collagens, Fibrosis, Mouse models
CCR10+ epithelial cells are abundantly detected in lungs from IPF patients, where these cells promote IPF lung fibroblast invasion, collagen 1 secretion, and lung fibrosis in humanized mice.
Introduction
Idiopathic pulmonary fibrosis (IPF) is the most common clinical form of interstitial lung disease (ILD), with poor prognosis and a median survival of 3–5 years after diagnosis. This devastating disease is characterized histologically by the presence of usual interstitial pneumonia (UIP), containing fibroblastic foci, which are believed to be the site of active tissue remodeling. The incidence of IPF ranges from 14.6–28.8 cases per 100,000 persons (1–3) and its prevalence increases with age. The fibrotic triggers in IPF are unknown but it is speculated that persistent lung injury leads to alveolar epithelial cell injury and death, and subsequent epithelial progenitor exhaustion and/or dysfunction lead to disrepair mechanism(s) that ablate the alveolus. Currently, 2 therapeutics have been approved for the treatment of IPF, Esbriet (4, 5) and OFEV (6, 7), both of which have been shown to slow down but not halt disease progression, and neither drugs promote disease regression and lung regeneration. Thus, there is significant effort underway to develop next-generation therapeutics that might further slow or halt progressive lung remodeling in these patients.
Due to its idiopathic nature, several epidemiological studies have assessed potential links between lifestyle, environmental contaminants and this disease, leading to a consensus that epithelial injury and subsequent aberrant and chronic activation of stromal cells may be important contributing factors to the progressive remodeling observed in IPF lungs. Utilizing mouse models of this disease, several studies have identified PDGFR-expressing interstitial fibroblasts/lipofibroblasts and pericytes as the major contributors to collagen-expressing fibroblasts and myofibroblasts in fibrotic mouse lungs (8–11). Further, other studies have observed an expansion of KRT5+ cells into the alveolar parenchyma of bleomycin-injured or influenza-infected mouse lungs (12, 13), with one study suggesting that these cells may be distinct from airway basal and club cells (12). Consistently, several studies have reported an expansion of KRT5+ and SCGB1A1+ epithelial cells in fibrotic IPF lungs (14–16); however, the role of these epithelial cells in the relentless and progressive lung remodeling observed in this disease remains elusive.
Herein, we show that there is a significant increase in the percentage of EpCAM+CCR10+ epithelial cells in IPF relative to normal explanted lung tissues. Utilizing a humanized mouse model of IPF (in which IPF structural and immune cells are intravenously administered into immunocompromised NSG mice induce fibrotic lung remodeling; see ref. 17), the numbers of engrafted EpCAM+CCR10+ human epithelial cells in NSG lungs significantly correlated with lung remodeling in the humanized mice. Utilizing several in vitro culture techniques, airway KRT5+ and SCGB1A1+ epithelial cells from explanted normal and IPF lung tissues were expanded and characterized. Conditioned supernatants (CSs) from cultured IPF epithelial cells enhanced IPF lung fibroblast invasion and collagen 1 secretion. Single-cell transcriptomic analysis of EpCAM+CCR10+ epithelial cells identified distinct KRT5+ and SCGB1A1+ proinflammatory and profibrotic epithelial cell populations. Intravenous administration of cultured IPF (CCR10+ epithelial cell enriched), but not normal (containing significantly less CCR10+ cells than IPF), epithelial cells induced lung remodeling in NSG mice. Importantly, the numbers of CCR10+ IPF but not normal epithelial cells significantly correlated with hydroxyproline concentration in the remodeled mouse lung tissues. Further, the majority of epithelial cells highly expressing CCR10 also express the receptor tyrosine kinase EphA3, and activation of Ephrin A signaling promoted CCR10 expression on the same cells. Finally, intravenous administration of EphA3+ epithelial cells, that highly express CCR10 transcript (i.e., CCR10hi), promoted more consistent lung remodeling in humanized NSG mice relative to EphA3– epithelial cells, expressing low levels of CCR10 transcript (i.e., CCR10lo), from the same explanted IPF lung. These studies suggest that CCR10hi epithelial cells promote lung fibrosis in IPF and that targeting profibrotic mechanisms elaborated by these cells may be beneficial in this disease.
Results
Abundance of EpCAM+CCR10+ epithelial cells in explanted IPF lungs that correlate with lung remodeling in humanized NSG mice.
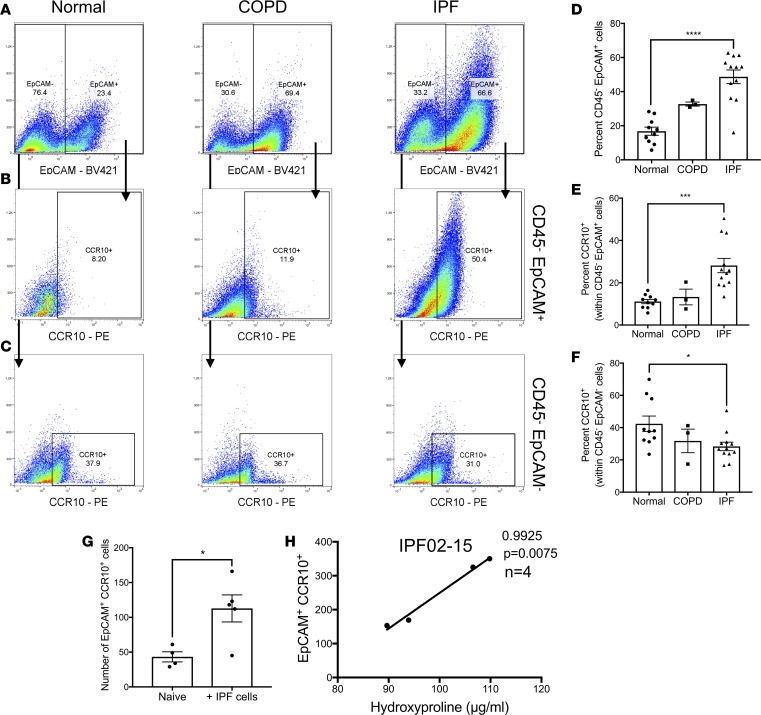
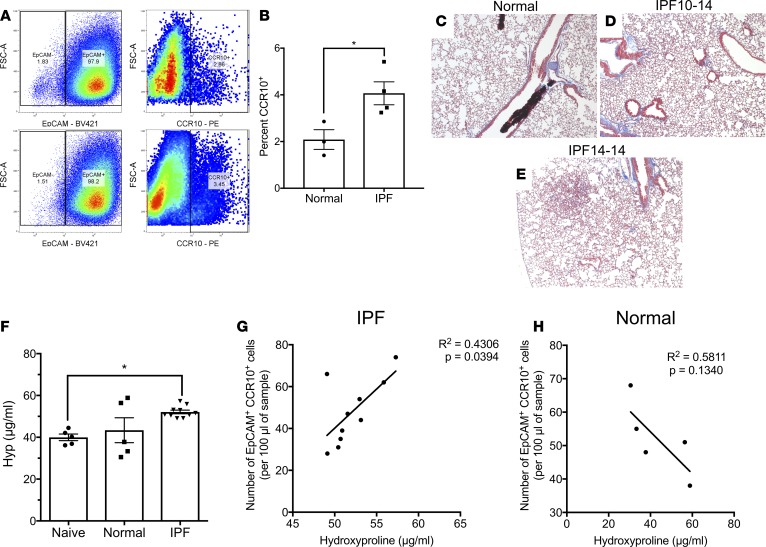
We have recently shown that CCR10 is highly expressed in progressive IPF patients’ diagnostic biopsies and on structural cells from IPF, but not normal lung explants (18). To further assess the expression of CCR10 on structural cells, normal, COPD, and IPF explanted lung cellular suspensions were stained with fluorescently conjugated anti-CD45, -EpCAM, and -CCR10 antibodies and flow cytometrically analyzed. There was significant increase in the percentage of CD45–EpCAM+ cells and CD45–EpCAM+ cells expressing CCR10 in IPF compared with normal lung explants (Figure 1, A and B and quantified in Figure 1, D and E). Further, the percentage of EpCAM–CD45–CCR10+ cells was significantly reduced in IPF relative to normal lung explants (Figure 1C and quantified in Figure 1F). These results suggest that CCR10-expressing epithelial cells are abundantly detected in IPF lung explants.
Figure 1. EpCAM+CCR10+ epithelial cells are abundant in IPF lung explants and their numbers correlated to lung remodeling in humanized NSG mice.
(A–C) Normal, COPD, and IPF lung explant cellular suspensions were stained with fluorescently conjugated anti-CD45, anti-EpCAM, and anti-CCR10 antibodies and analyzed by flow cytometry. Shown are representative dot plots from normal (left, n = 10), COPD (middle, n = 3), and IPF (right, n = 12) lung explants depicting CD45–EpCAM+ (A), CD45–EpCAM+CCR10+ (B) and CD45–EpCAM–CCR10+ (C) cells. (D–F) Shown is the percentage of CD45–EpCAM+ (D), CCR10+ cells within CD45–EpCAM+ (E) and CD45–EpCAM– (F) cells in normal, COPD, and IPF lung explants. Data shown are the mean ± SEM. *P ≤ 0.05; ***P ≤ 0.001; ****P ≤ 0.0001 via 1-way ANOVA corrected with Dunnett’s test. NSG-GFP or NSG mice were intravenously administered with IPF lung explant cells; 35 days after cellular administration, lung cellular suspensions were analyzed by flow cytometry and hydroxyproline concentration was quantified from lung homogenates. (G) Depicted is the mean number of GFP–EpCAM+CCR10+ cells in the lungs of naive and IPF humanized NSG-GFP mice. Data shown are the mean ± SEM. *P ≤ 0.05 via unpaired Mann-Whitney 2-tailed nonparametric test. (H) Depicted is a correlation analyses of hydroxyproline concentration and number of human CD45–EpCAM+CCR10+ cells in IgG-treated NSG lungs after 35 days of IPF cell administration. n = 4–5/group. P values indicated. IPF, idiopathic pulmonary fibrosis; NSG, NOD Cg-PrkdcSCID IL2rgTm1wilSzi; NSG-GFP, NOD.Cg-PrkdcSCID IL2rgtm1Wil Tg (CAG-EGFP) 10sb/SzJ.
To assess the potential role of CCR10-expressing epithelial cells in lung remodeling, a humanized NSG model of IPF was utilized (17). Thirty-five days after IPF cellular administration in NSG mice transgenically expressing GFP (NSG-GFP), GFP–EpCAM+CCR10+ human cells engrafted in the lungs of NSG mice as shown by the significant increase in the numbers of these cells in humanized relative to naive, nonhumanized, NSG lungs (Figure 1G). Further, there was a significant positive correlation between the numbers of engrafted CD45–EpCAM+CCR10+ cells and hydroxyproline concentration in humanized NSG lungs (Figure 1H). These results suggest that IPF CCR10+ epithelial cells engraft and promote fibrotic lung remodeling in humanized NSG mice.
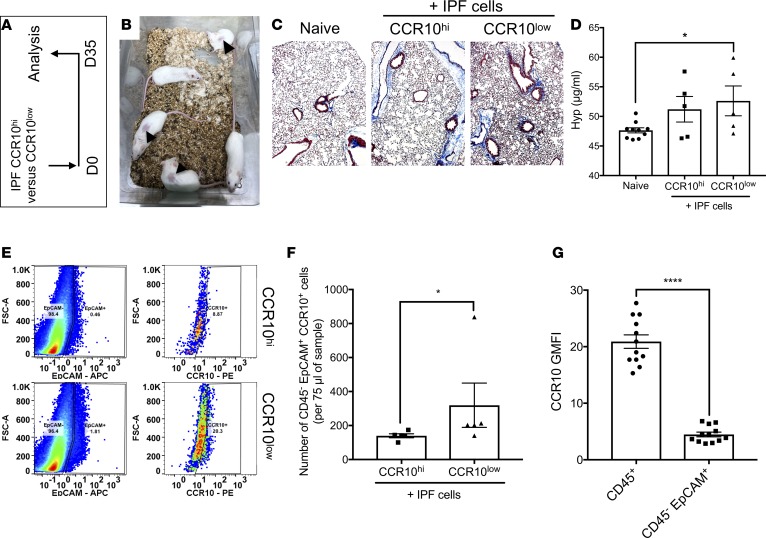
To assess potential heterogeneity of CCR10-expressing cells in IPF lungs, IPF lung explant cells were magnetically enriched using anti-CCR10 antibodies. Quantitative PCR (qPCR) analysis of the sorted and nonsorted cells showed that both groups expressed CCR10 transcript, with the sorted group having significantly higher CCR10 transcript expression (not shown). Thus, sorted and nonsorted cells were designated as CCR10hi and CCR10lo, respectively, based on transcript expression. To assess the role of IPF CCR10hi and CCR10lo cells in the promotion of lung remodeling in vivo, 2.5 × 105 sorted cells were i.v. administered into NSG mice (Figure 2A). Thirty-one days after cellular administration, there were visible skin lesions in the CCR10hi humanized NSG group (Figure 2B). Given the severity of these lesions, the experiment was terminated after 35 days, when there was more consistent histological evidence for mild interstitial consolidation and focal regions of remodeling in the lungs of NSG mice humanized with IPF CCR10lo (Figure 2C, right), relative to CCR10hi or naive NSG groups (Figure 2C, middle and left, respectively). At this time, lung hydroxyproline was significantly elevated in CCR10lo, but not CCR10hi, humanized relative to the naive NSG groups (Figure 2D). Finally, flow cytometric analysis showed a significant increase in the numbers of CD45–EpCAM+CCR10+ cells in CCR10lo compared with CCR10hi humanized NSG groups (Figure 2E, and quantified in Figure 2F). Consistently, IPF CD45–EpCAM+ epithelial cells showed significantly lower geometric mean fluorescence intensity (GMFI) staining for CCR10 compared with CD45+ immune cells. These results suggest that epithelial cells express less CCR10 protein on their cell surface relative to immune cells. Further, the numbers of CCR10+ epithelial cells were significantly higher in the lungs of IPF CCR10lo cell–humanized NSG mice, thus leading us to speculate that CCR10+ epithelial cells in these samples may have promoted lung remodeling in the humanized mice.
Figure 2. CCR10lo-expressing epithelial cells engraft and promote fibrosis in the lungs of humanized NSG mice.
(A) Magnetically sorted IPF CCR10hi and CCR10lo cells were intravenously administered into NSG mice (0.25 million cells/mouse) and after 35 days, the mice were sacrificed, their lungs were collected for histological, biochemical, and flow cytometric analyses. (B) Shown is an image of IPF CCR10hi-humanized NSG mice, 31 days after cellular administration. (C) Shown are representative images of lung tissues, from naive (left), IPF CCR10hi-humanized (middle), and IPF CCR10lo-humanized (right) NSG groups taken at ×100 magnification. (D) Depicted is the hydroxyproline concentration in the lungs of naive, nonhumanized, or humanized NSG mice that received IPF CCR10hi or CCR10lo cells. n = 5–10 mice/group. Data shown are the mean ± SEM. ns, not significant; *P ≤ 0.05 via 1-way ANOVA corrected for multiple comparison via Dunnett’s test. (E) CCR10hi or CCR10lo humanized NSG mouse lung suspensions were analyzed by flow cytometry for human CD45, EpCAM, and/or CCR10 expression. Depicted are representative dot plots showing EpCAM+ cells (left) and the percentage of EpCAM+ cells expressing cell surface CCR10 (right) in IPF CCR10hi (top) and CCR10lo (bottom) humanized NSG lungs. (F) Shown is the average number of EpCAM+CCR10+ cells in IPF CCR10hi and CCR10lo humanized NSG mice. n = 5/group. Data shown are the mean ± SEM. *P ≤ 0.05 via 2-tailed Mann-Whitney nonparametric test. (G) IPF lung explant cells were stained with anti-CD45, anti-EpCAM, and anti-CCR10 antibodies. Shown is the mean GMFI for CCR10 staining on CD45+ and CD45–EpCAM+ cells. n = 12/group. Data shown are the mean ± SEM. ****P ≤ 0.0001 via 2-tailed Mann-Whitney nonparametric test. IPF, idiopathic pulmonary fibrosis; NSG, NOD Cg-PrkdcSCID IL2rgTm1wilSzi.
Given the histocompatibility mismatch between the human and murine cells, it is possible that human CCR10+ T cells may induce injury leading to a remodeling response involving CCR10+ epithelial cells. Thus, to assess T cell independent roles of CCR10+ epithelial cells in lung remodeling using this humanized model, FACS-isolated CD3–CCR10+ cells were intravenously administered into NSG mice (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.122211DS1). Thirty-five days after cellular administration, the number of CD45–EpCAM+CCR10+ cells positively correlated with hydroxyproline levels in humanized NSG lungs (Supplemental Figure 1B). These results suggest that CCR10-expressing epithelial cells may be enriched in CCR10lo samples that induced lung remodeling in humanized NSG mice. Further, the number of these CCR10+ epithelial cells positively correlated with the concentration of hydroxyproline in remodeled NSG lungs humanized with a mixture of immune and nonimmune IPF lung explant cells.
In vitro culture, expansion and characterization of normal and IPF explanted lung–derived airway epithelial cells.
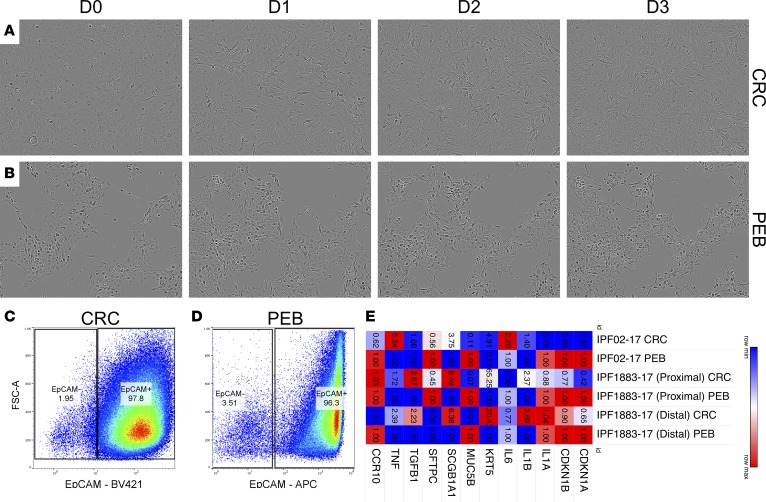
Given the abundance of CCR10-expressing epithelial cells in IPF lungs, in vitro culture optimization and characterization of normal and IPF lung epithelial cells was undertaken. Normal and IPF lung epithelial cells were cultured as described in the Methods section using a modified conditionally reprogrammed cell (CRC) culture technique (19, 20) or using PneumaCult-Ex Plus Medium containing 10 μM Y27632 (i.e., PEB medium). After approximately 2–3 weeks, epithelial colonies were apparent in these cultures (Figure 3, A and B), in which epithelial cells cultured in CRC medium showed a hyperplastic and less cuboidal structure (Figure 3A and Supplemental Video 1) compared with cells cultured in PEB medium (Figure 3B and Supplemental Video 2) over a duration of 3 days. Epithelial cells cultured in CRC medium showed a partial reversion to a cuboidal-like structure after the replacement of CRC medium with PEB medium (Supplemental Video 3). Confluence of epithelial cells and the number of passages prior to growth arrest were generally higher under CRC conditions. Flow cytometric analysis showed that the majority of CRC or PEB cultured cells highly expressed EpCAM (96%–98%; Figure 3, C and D). Transcriptomic analysis showed that both normal and IPF lung epithelial cultures were enriched for basal (KRT5) and club cell (SCGB1A1) markers; however, these cells also expressed lower levels of MUC5B and SFTPC transcripts (Supplemental Figure 2). Cells cultured in CRC medium expressed lower levels of CDKN1A, CDKN1B, MUC5B, SFTPC and, less consistently, CCR10 transcripts relative to PEB-medium-cultured cells (Figure 3E). Finally, CRC-cultured cells expressed higher levels of IL1B, SCGB1A1, TNF, KRT5 and, less consistently, TGFB1 transcripts relative to cells cultured in PEB medium (Figure 3E). These results suggest that primary airway epithelial cells can be cultured using either CRC or PEB media, where cells cultured under CRC conditions appear to proliferate more rapidly and express less cell cycle inhibitory proteins.
Figure 3. Characterization of in vitro–cultured airway epithelial cells.
Normal and IPF explant lung cellular suspensions were cultured in CRC or PEB media. (A and B) Depicted are representative images taken at ×100 magnification showing the morphology of cultured epithelial cells in CRC (A) or PEB (B) media over the duration of 3 days. (C and D) Cultured epithelial cells were dissociated using Accutase cell detachment solution, stained with fluorescently conjugated anti-EpCAM antibodies, and analyzed by flow cytometry. Shown are representative dot plots of EpCAM+ cells cultured in CRC (C) or PEB (D) media. n = 7 CRC; n = 2 PEB medium. (E) RNA was extracted from PEB- or CRC-cultured epithelial cells and quantitative PCR analysis was performed for markers of epithelial cell lineages, quiescence/senescence, fibrosis, and inflammation. Depicted is a Morpheus-generated heatmap (broadinstitute.org) showing fold change of expression in CRC- relative to PEB-cultured epithelial cells from 2 distal lung tissues (IPF02-17 and IPF 1883-17 distal) and 1 proximal tissue (IPF1883-17 proximal) derived from 2 explanted IPF lungs. Colors depict the relative expression of each transcript, where higher or lower expression is depicted in red or blue, respectively. The numbers in each quadrant indicate the fold change of expression. CRC, conditionally reprogrammed cell medium; PEB, PneumaCult-Ex Plus Medium; IPF, idiopathic pulmonary fibrosis.
CRC culture conditions were reported to require irradiated feeder cells for optimal cellular growth (19, 20). Given that irradiation of feeder cells induces their senescence (21), we hypothesized that factors generated by senescent fibroblasts may mediate cell survival and/or proliferation of the cultured epithelial cells. Further, given the expansion of airway epithelial cells in IPF lungs (14–16), we also hypothesized that cytokines abundantly expressed in IPF lung tissues may promote the survival and/or proliferation of these cells. To test these hypotheses, lung epithelial cells were cultured in DMEM containing 15% FBS, antibiotics, and L-glutamine in the presence or absence of 10 μM Y27632 or various components that were previously reported to be generated by lung fibroblasts and/or senescent lung fibroblasts (i.e., hypomethylated CpG-DNA [CpG]; ref. 22, or IL-1β/IL-18; refs. 23–25) or abundantly detected in IPF lung tissues (i.e., IL-4/IL-13; refs. 26–32). As a control, epithelial cells were cultured in CSs from senescent lung fibroblasts (Sen C/S) in the presence or absence of 10 μM Y27632. Kinetic live cell imaging analysis using an Incucyte Zoom system showed that epithelial cells cultured in the absence of senescent medium or Y27632 progressively died in culture, as evident by their decreased confluence overtime (Supplemental Figure 3, A–C, black line). Similar findings were also observed with cells cultured in PEB medium in the absence of Y27632 (Supplemental Video 4). Senescent medium, CpG, and IL-13 consistently improved the survival (Supplemental Figure 3, A–C and G–I) and IL-4, IL-1β, or IL-18 also improved the survival, albeit less consistently, (Supplemental Figure 3, G–I and M–O) of untreated epithelial cells. Cells cultured in the presence of 10 μM Y27632 proliferated over the duration of the experiment (Supplemental Figure 3, D–F, black line and Supplemental Videos 1 and 2) and this proliferative response was promoted by the addition of senescent medium, CpG, or IL-13 (Supplemental Figure 3, D–F and J–L). Further, IL-4, IL-1β, and IL-18 less consistently promoted the proliferation of epithelial cells cultured in the presence of 10 μM Y27632 (Supplemental Figure 3, J–L and P–R). These results suggest that Y27632 is required for the proliferation of epithelial cells and senescent medium, and either hypomethylated-CpG DNA or IL-13 further enhance this proliferative response. Thus, all subsequent characterization and functional studies were performed in the presence of 10 μM Y27632 in Sen C/S or PEB medium.
IPF but not normal lung airway epithelial cells’ CSs induce IPF lung fibroblast invasion and collagen 1 secretion.
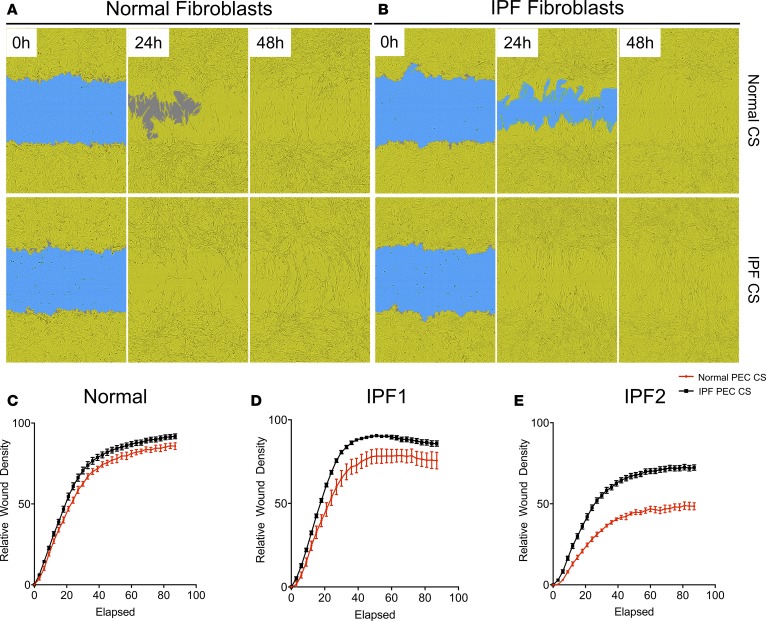
To assess the modulation of lung fibroblasts by cultured normal and IPF epithelial cells, lung epithelial cells were cultured in PEB medium for 24 hours, after which their CSs were collected and utilized to stimulate lung fibroblasts. As controls, CSs were collected from empty cell-free wells treated in a similar manner (cell-free). IPF epithelial cell CS promoted invasive wound healing of IPF (Figure 4B and quantified in Figure 4, D and E) and, to a lesser extent, normal lung fibroblasts (Figure 4A and quantified in Figure 4C) relative to normal epithelial cell CS. Further, both normal and IPF epithelial cell CS induced a modest elevation of collagen 1 protein in normal lung fibroblasts (Supplemental Figure 4A); however, IPF epithelial cell CS more consistently induced a significant elevation in collagen 1 protein secretion from IPF lung fibroblasts (Supplemental Figure 4, B and C). These results suggest that IPF epithelial cells promote lung fibroblast invasion and collagen 1 secretion.
Figure 4. IPF cultured epithelial cell–conditioned supernatants promote lung fibroblast invasion.
Normal and IPF explant suspensions were cultured in PEB medium until epithelial cell colonies were apparent. After 2–3 passages, cells were cultured in 6-well plates for 24 hours, after which culture supernatants (CSs) were collected and frozen at –20°C. Lung fibroblasts were then plated into 96-well plates, scratched using a WoundMaker, and treated with normal or IPF epithelial cell CS mixed with Matrigel. Lung fibroblast invasion was monitored using an Incucyte Zoom live cell imager. (A and B) Depicted are representative images showing the closure of wounded areas (blue) by normal (n = 1; A) or IPF (n = 2; B) lung fibroblasts (yellow) after treatment with normal (n = 2; top) or IPF (n = 4; bottom) epithelial cell CS over the span of 48 hours. (C–E) Depicted is the average wound closure (relative to the initial wound) over 90 hours of normal (n = 1) or IPF (n = 2) fibroblasts treated with 2 normal (red) or 4 IPF (black) PEC CSs. Data shown are the mean ± SEM. PEB, PneumaCult-Ex Plus Medium + 10 µM Y27632; PEC, pulmonary epithelial cells; IPF, idiopathic pulmonary fibrosis.
Single-cell transcriptomic analysis of CCR10+ IPF epithelial cells.
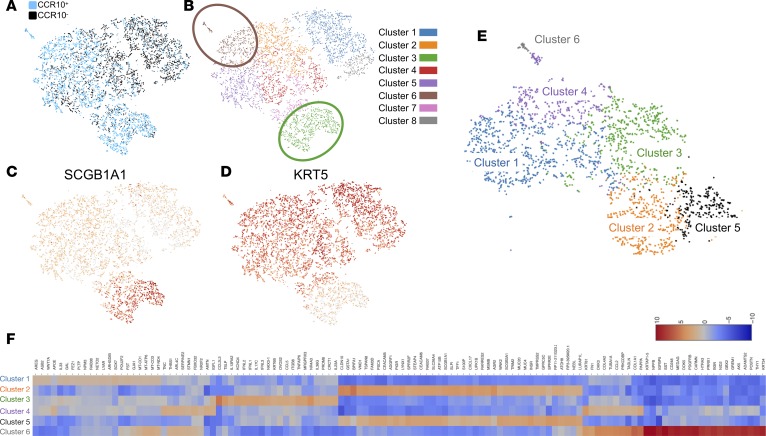
Given the correlation between the numbers of CCR10+ epithelial cells and hydroxyproline concentration in IPF explant-cell-humanized NSG lungs and the promotion of lung fibroblast invasion and collagen 1 secretion by cultured IPF epithelial cells, we hypothesized that CCR10+ epithelial cells may be inherently profibrotic and thus their abundance in IPF samples might contribute to the observed fibroblast activation and invasive responses in vitro and remodeling responses in vivo. To elucidate potential profibrotic mechanisms elaborated by CCR10+ IPF epithelial cells and heterogeneity within these cells, IPF epithelial cells were generated by culturing lung explant suspensions in CRC medium (to promote rapid cell expansion; as shown in Figure 3A) and then transferred into PEB medium and incubated overnight (to promote CCR10 expression; as shown in Figure 3E). Epithelial cells were then dissociated and stained with anti-EpCAM and -CCR10 antibodies. EpCAM+CCR10+ and EpCAM+CCR10– epithelial cells were then FACS isolated and 3,575 CCR10+ and 3,863 CCR10– epithelial cells were captured and single-cell RNA sequenced using the 10× Genomics platform. RNA-seq analysis yielded 88,840 (5,704 median genes/cell) and 75,271 (5,342 median genes/cell) mean reads per CCR10+ and CCR10– epithelial cells, respectively. The resulting data were then aggregated, where graph-based analysis showed that CCR10+ epithelial cells were distinct from CCR10– epithelial cells (Figure 5A, blue versus black, respectively). There were 2 clusters that were distinctly enriched for CCR10+ cells, graph-based clusters 3 and 6 (Figure 5B, brown and green clusters). Further, graph-based cluster 5 (Figure 5B, purple) was also enriched for CCR10+ cells but contained a few CCR10– epithelial cells. Transcript analysis for epithelial lineage markers (enriched using our culture techniques [as shown in Supplemental Figure 2]) showed that cells from cluster 3 highly expressed the club cell marker, SCGB1A1 (Figure 5C), and cells from cluster 6 highly expressed the basal cell marker KRT5 (Figure 5D). K-mean cluster analysis of CCR10+ cells subclustered cells from graph-based cluster 6 (Figure 5B, brown) into 2 clusters, K-mean clusters 4 and 6 (Figure 5E, purple and gray); showed 2 additional KRT5-expressing clusters (K-mean clusters 1 and 3; Figure 5E, blue and green) and 2 SCGB1A1-expressing clusters (K-mean clusters 2 and 5; Figure 5E, orange and black). Differentially expressed transcriptomic analysis on these K-mean clusters showed that cells from K-mean cluster 6 expressed high levels of stromal cell markers, including COL1A1, COL1A2, COL3A1, FN1, PDGFRB, and THY1 (Figure 5F, gray and Supplemental Table 1). Further, these cells expressed a few keratin proteins, including KRT34 and KRT81. Cells from K-mean cluster 4 expressed fewer COL1A1, COL1A2, COL3A1, and FN1 transcripts, expressed similar levels of KRT81 transcripts relative to K-mean cluster 6, and were specifically enriched for TNC, FBXO32, HOXB5, ARL4C, THBS1, FN1, CDH2, AMTN, and SERPINE2 transcripts relative to the other clusters (Figure 5F, purple and Supplemental Table 1). Cells from K-mean cluster 1 (graph-based cluster 5; enriched for CCR10+ cells but also contained CCR10– cells; Figure 5, A, B, and E) were enriched for SOX7, LYAR, GAL, FST, WNT7A, POU2F2, AREG, PLTP, GJA1, and IL33 transcripts relative to the other clusters (Figure 5F, blue). SCGB1A1+ cells from K-mean clusters 2 and 5 expressed similar overall transcripts but were specifically enriched for AGR2, CLDN10, CXCL17, GSTA1, SLPI, TSPAN8, S100P, MSMB, SCGB1A1, and WFDC2 in K-mean cluster 2 and INSR, MUC4, WNK2, NEBL, CECAM6, ATOH8, TRIM2, TMPRSS2, GPRC5C, and STEAP4 in K-mean cluster 5 relative to the other clusters (Figure 5F, orange and black, respectively; Supplemental Table 1). These results suggest that CCR10+ epithelial cells contain distinct epithelial cell populations relative to CCR10– cells, and several populations in the former group express proinflammatory and profibrotic transcripts.
Figure 5. Single-cell RNA-seq analysis of EpCAM+CCR10+ and EpCAM+CCR10– epithelial cells shows unique epithelial cell populations enriched by CCR10.
IPF epithelial cells were generated in CRC medium and then incubated in PEB medium overnight to promote CCR10 expression. Cells were then dissociated using Accutase and stained with fluorescently conjugated anti-EpCAM and -CCR10 antibodies. EpCAM+CCR10+ and EpCAM+CCR10– epithelial cells were FACS isolated and single-cell RNA-seq analysis was performed using the 10× Genomics platform. (A and B) Depicted are images showing CCR10+ versus CCR10– sequenced epithelial cells (A) and graph-based clustering of sequenced epithelial cells (B). Unique cell populations enriched in CCR10+ (blue) relative to CCR10– (black) epithelial cell populations are highlighted (brown and green circles). (C and D) Depicted are images showing SCGB1A1-expressing (C) or KRT5-expressing (D) epithelial cells. (E and F) Shown is K-mean cluster analysis of CCR10+ epithelial cells (E) and a heatmap of differentially expressed transcripts in cells from each K-mean cluster (F). CRC, conditionally reprogrammed cell medium; PEB, PneumaCult-Ex Plus Medium + 10 µM Y27632; FACS, fluorescence-activated cell sorting; IPF, idiopathic pulmonary fibrosis.
To further assess pathways enriched in CCR10+ cells, population transcriptomic assessment was performed. Single-cell transcriptomic datasets were imported into SeqGeq (FlowJo, LLC) and the samples were concentrated. PCA and tSNE analysis were performed to generate statistically distinct population of cells (Supplemental Figure 5A, CCR10+ cells in red and CCR10– cells in blue). Consistent with our graph-based analysis (Figure 5, A–D), CCR10+ cells were enriched in SCGB1A1-expressing (Supplemental Figure 5B) and KRT5-expressing (Supplemental Figure 5C) cell populations. To perform population analysis, KRT5+CCR10+, KRT5+CCR10–, and SCGB1A1+CCR10+ cells were gated (Supplemental Figure 5D) and significantly differentially expressed genes in these CCR10+ versus CCR10– cell populations were determined (Supplemental Figure 5, E–G) and uploaded onto Ingenuity IPA (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis) for pathway and upstream analysis. Ingenuity canonical pathway analysis of transcripts that were enriched in KRT5+CCR10+ versus KRT5+CCR10– (Supplemental Figure 5E) cells showed enrichment for transcripts involved in oxidative phosphorylation, mitochondrial dysfunction, phagosome maturation, molecular mechanisms of cancer, death receptor signaling, and interferon signaling Ingenuity canonical pathways (Supplemental Table 2). Ingenuity upstream regulator analysis predicted the activation of several inflammatory and profibrotic upstream regulators including IFNA2, TNF, HNF4A, IFNG, TP53, NFKB (complex), OSM, IRF7, TGFB1, and STAT1 (Supplemental Table 3) in KRT5+CCR10+ versus KRT5+CCR10– cells. Ingenuity canonical pathway analysis of SCGB1A1+CCR10+ cells versus KRT5+CCR10– cells (Supplemental Figure 5F) showed an enrichment of transcripts involved in oxidative phosphorylation, mitochondrial dysfunction, phagosome maturation and Interferon signaling pathways in the former cell populations (Supplemental Table 4). Ingenuity upstream regulator analysis also predicted the activation of several inflammatory and profibrotic regulators including, HNF4A, IFNG, IFNA, TNF, IFNA2, STAT1, OSM, STAT5A, IRF7, IL1B, and IRF1 in SCGB1A1+CCR10+ relative to KRT5+CCR10– epithelial cells (Supplemental Table 5). Finally, Ingenuity canonical pathway analysis of CCR10-expressing SCGB1A1+ versus KRT5+ epithelial cells (Supplemental Figure 5G) showed that SCGB1A1+ cells were enriched for transcripts involved in mitochondrial dysfunction, NRF2-mediated oxidative stress responses, aryl hydrocarbon receptor signaling, oxidative phosphorylation, unfolded protein response, and ER stress pathways (Supplemental Table 6). Ingenuity upstream regulator analysis showed that the inflammatory and profibrotic upstream regulators, IFNG, TNF, STAT5A, HNF4A, STAT1, IFNA2, and IL1B were predicted to be activated in CCR10-expressing SCGB1A1+ versus KRT5+ cells (Supplemental Table 7). These results suggest that CCR10+ epithelial cells are enriched for proinflammatory and profibrotic transcripts relative to CCR10– cells and SCGB1A1+CCR10+ cells were enriched for transcripts involved in epithelial stress responses relative to KRT5+CCR10+ cells.
IPF but not normal epithelial cells induce lung fibrosis in humanized NSG mice.
Flow cytometric analysis of cultured epithelial cells showed that there was significant increase in the percentage of CCR10+ epithelial cells in IPF relative to normal epithelial cultures (Figure 6, A and B). To assess the function of these epithelial cells in vivo, 5.0 × 105 normal or IPF epithelial cells were intravenously administered into NSG mice, and 63 days later the mice were sacrificed, and lungs were collected for flow cytometric assessment of EpCAM+CCR10+ human epithelial cells, histological, and biochemical analysis for collagen deposition. Sixty-three days after epithelial cell administration, there was evidence of interstitial thickening in NSG mice that received cultured IPF (Figure 6, D and E), and to a lesser extent, normal (Figure 6C) lung epithelial cells, as assessed histologically via Masson’s trichrome staining. Most importantly, the lungs of NSG mice that received cultured IPF epithelial cells more consistently showed a significant elevation in hydroxyproline concentration compared with lungs of NSG mice that received normal epithelial cells (Figure 6F). Finally, flow cytometric quantification of human-CCR10-expressing epithelial cells in both groups of humanized NSG mice showed that there was a significant positive correlation between the number of CD45–EpCAM+CCR10+ epithelial cells and hydroxyproline concentration in the lungs of NSG mice that received IPF (Figure 6G) but not normal (Figure 6H) epithelial cells. Together, these results demonstrate that epithelial cells from IPF, but not normal lungs, induce consistent lung remodeling in NSG mice at day 63 after intravenous administration and this remodeling response positively correlated with the number of engrafting IPF CCR10+ epithelial cells.
Figure 6. IPF epithelial cells induce lung remodeling in NSG mice.
(A) CRC-cultured normal (n = 3) and IPF (n = 4) epithelial cells were dissociated using Accutase solution, stained with fluorescently conjugated anti-EpCAM and -CCR10 antibodies and analyzed by flow cytometry. Shown are representative dot plots of EpCAM+ cells (left) and CCR10+ cells within the EpCAM+ cells (right) from normal (n = 3; top) and IPF (n = 4; bottom) epithelial cell cultures. (B) Shown is the percentage of CCR10+ epithelial cells from normal and IPF epithelial cell cultures. Data shown are the mean ± SEM. n = 3–4/group. *P ≤ 0.05 via unpaired parametric t test. (C–E) Normal and IPF culture–expanded epithelial cells were injected intravenously into NSG mice and allowed to engraft for 63 days, after which the mice were sacrificed, and their lungs were analyzed for remodeling. Representative Masson’s trichrome staining taken at ×50 magnification of NSG mouse lungs, 63 days after normal (C) and IPF (D and E) epithelial cell administration. (F) Depicted is the average hydroxyproline content in the superior and middle lobes in unchallenged, naive, and epithelial cell–challenged NSG mice. Data shown are the mean ± SEM. n = 5 from 1 normal donor lung; n = 10 from 2 IPF patients’ lungs. *P ≤ 0.05 via 1-way ANOVA corrected with Dunnett’s test. (G and H) Cellular suspensions from humanized NSG lung tissues were generated and analyzed by flow cytometry for human EpCAM+CCR10+ cells. Depicted are correlation analyses of hydroxyproline concentration and number of human CD45–EpCAM+CCR10+ cells in NSG lungs humanized with IPF (G) or normal (H) epithelial cells at day 63 after cell injection. n = 5 from 1 normal donor lung; n = 10 from 2 IPF patients’ lungs. P values indicated. IPF, idiopathic pulmonary fibrosis; NSG, NOD Cg-PrkdcSCID IL2rgTm1wilSzi; Hyp, hydroxyproline.
EphA3 is expressed by epithelial cells lining micro-honeycombing cysts, where its signaling induces CCR10 expression in IPF but not normal epithelial cells.
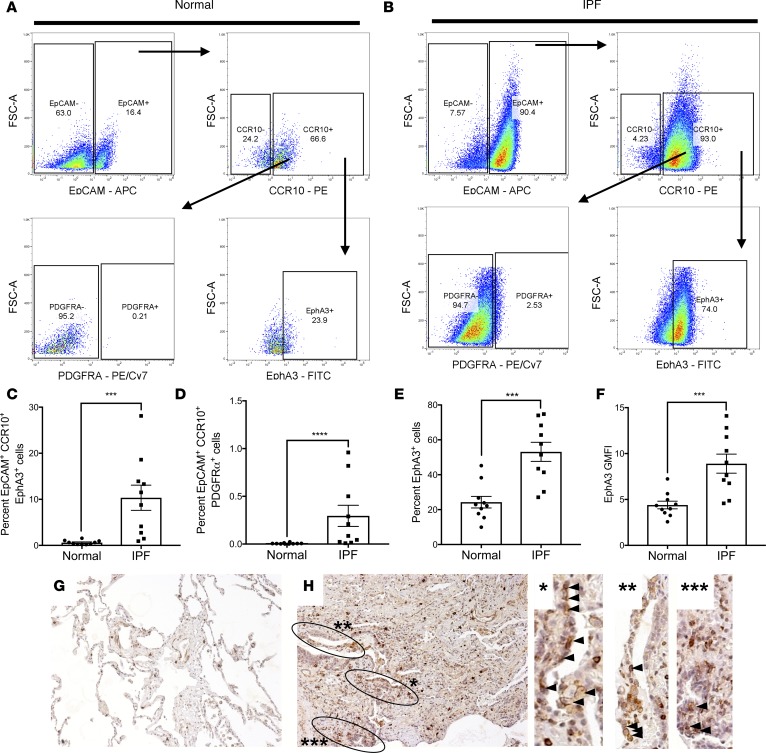
We have recently shown that a subset of CCR10+ structural cells coexpress the receptor tyrosine kinase EphA3 (ref. 18 and manuscript in revision), a putative pulmonary mesenchymal marker (33). Given the expression of a few mesenchymal markers in single-cell RNA-sequenced KRT5+CCR10+ epithelial cells (as shown in Figure 5, E and F), the expression of EphA3 on CCR10+ epithelial cells was assessed. Flow cytometric analysis of CCR10+ epithelial cells in normal and IPF lung explant cells showed that a subset of normal CCR10+ epithelial cells expressed cell-surface EphA3 but not PDGFRα protein (another mesenchymal marker in the lung; refs. 8, 34) (Figure 7A and quantified in Figure 7, C and D); however, there was a significant increase of the percentage of EpCAM+CCR10+EphA3+ (Figure 7B and quantified in Figure 7C) and EpCAM+CCR10+PDGFRα+ (Figure 7B and quantified in Figure 7D) epithelial cells in IPF relative to normal lungs. Further, there was a significant increase in the percentage of EphA3+ cells and EphA3 GMFI on CCR10-expressing epithelial cells in IPF relative to normal lung explants (Figure 7, E and F). To assess the relative expression of EphA3 on CCR10+ versus CCR10– epithelial cells in normal and IPF lung explants, flow cytometric analysis from normal and IPF lung explant cell samples comparing the 2 populations was performed. Both CCR10+ and CCR10– normal and IPF lung epithelial cells populations contained cells that stained for EphA3 (Supplemental Figure 6, A and B); however, in a subset of IPF patients, the majority of EphA3+ epithelial cells in IPF lung tissues coexpressed CCR10 (Supplemental Figure 6B and quantified in Supplemental Figure 6C). Further, both normal and IPF CCR10+ epithelial cells showed a significantly higher GMFI for EphA3, and IPF CCR10+ cells showed a significantly higher GMFI for EphA3 relative to normal CCR10+ cells (Supplemental Figure 6D). These results suggest that a subset of IPF patients’ CCR10-expressing epithelial cells abundantly expressed EphA3 protein, and that IPF CCR10+ epithelial cells expressed significantly higher levels of EphA3 protein relative to CCR10– and normal CCR10+ epithelial cells.
Figure 7. EphA3 is expressed by a subset of CCR10+ epithelial cells.
(A and B) Normal and IPF lung explant cellular suspensions were stained with fluorescently conjugated anti-CD45, -EpCAM, -CCR10, -EphA3 and/or -PDGFRα antibodies and analyzed by flow cytometry. Shown are representative dot plots from normal (A) or IPF (B) lung explants depicting CD45–EpCAM+ (top left), CD45–EpCAM+CCR10+ (top right), CD45–EpCAM+CCR10+PDGFRα+ (bottom left), and CD45–EpCAM+CCR10+EphA3+ (bottom right) cells. (C–F) Shown is the percentage of CD45–EpCAM+CCR10+EphA3+ (C) and CD45–EpCAM+CCR10+PDGFRα (D) cells in normal and IPF lung explants; and the percentage (E) and GMFI (F) of EphA3+ cells within the CD45–EpCAM+CCR10+ gate. n = 10 normal, n = 12 IPF. Data shown are the mean ± SEM. ***P ≤ 0.001; ****P ≤ 0.0001 via unpaired Mann-Whitney 2-tailed nonparametric test. (G and H) Representative IHC images stained for CCR10 (red) and EphA3 (brown) in normal lung (G), and IPF lung explants (H) taken at ×100 magnification. Dual-stained epithelial cells are highlighted in black circles and increased magnification images (H right, arrow heads) n = 5–8/group. IPF, idiopathic pulmonary fibrosis; GMFI, geometric mean fluorescence intensity.
To determine the potential localization of CCR10/EphA3-expressing epithelial cells in explanted lung tissues, dual-color immunohistochemical (IHC) analysis was performed. CCR10-immunopositive (red) and EphA3-immunopositive (brown) cells localized in the interstitium in normal lungs explants (Figure 7G) and in interstitial cells and cuboidal epithelial–like cells lining microcystic structures in IPF lung explants (Figure 7H, circled regions and arrow heads). Further analysis using polyclonal anti-EphA3 antibodies (specific C-terminal tail of EphA3, which does not bind to its ligands) showed that this protein is expressed by a subset of interstitial cells and in structures resembling microcapillaries in normal lung tissues (Supplemental Figure 7, A and B); however, this protein was predominantly expressed by immune-like interstitial and epithelial cells lining honeycombing cysts in IPF lung tissues (Supplemental Figure 7, C–F). Finally, IHC analysis of serial sections for KRT5, EphA3, and KRT81 (which was highly expressed in KRT5+CCR10+ epithelial cells; Figure 5F) showed that a subset of KRT5+ epithelial cells that localized adjacent to fibroblastic foci (Supplemental Figure 7G) stained with anti-EphA3 (Supplemental Figure 7H) and KRT81 (Supplemental Figure 7I) but not nonspecific IgG control antibodies (Supplemental Figure 7J). These results suggest that EphA3 is abundantly expressed by CCR10+ epithelial cells in IPF lungs and that a subset of these EphA3+ epithelial cells are localized adjacent to fibroblastic foci and honeycombing cysts in IPF lung tissues.
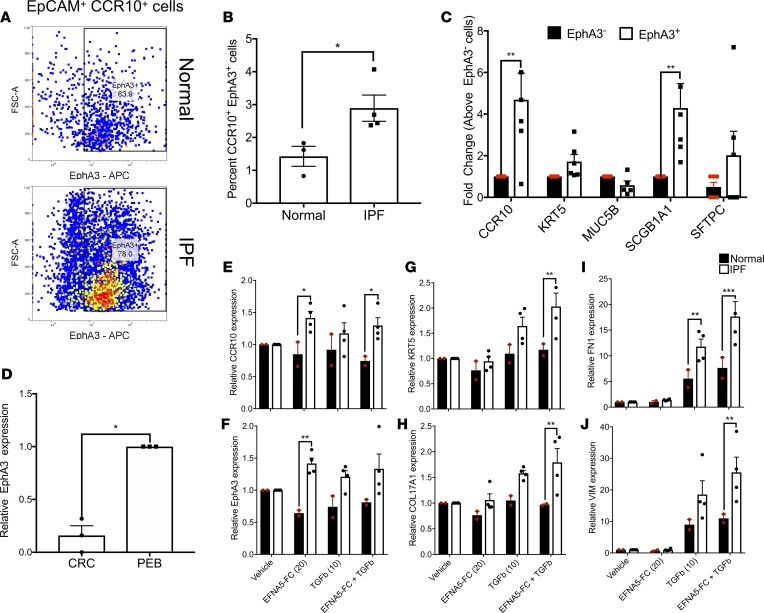
To assess the functional relevance of EphA3 on CCR10-expressing airway epithelial cells, in vitro analysis of this receptor in normal and IPF cultured epithelial cells was performed. Consistent with flow cytometric analysis of fresh lung explant derived epithelial cells, there was a significant increase in the percentage of CCR10+EphA3+ epithelial cells in IPF relative to normal epithelial cell cultures (Figure 8A and quantified in Figure 8B). Transcriptomic comparative analysis of magnetically sorted EphA3+ cells versus nonsorted EphA3– cells from epithelial cell cultures showed that the EphA3+ epithelial cells expressed significantly higher levels of CCR10 and SCGB1A1 transcripts relative to EphA3– epithelial cells (Figure 8C), which is consistent with the flow cytometric analysis of fresh and cultured epithelial cells (Figure 7, E and F and Figure 8, A and B). Further, transcriptomic analysis of CRC versus PEB medium–cultured IPF epithelial cells showed that PEB medium promoted the expression of EphA3 transcripts (Figure 8D). Thus, to assess the function of EphA3 signaling in IPF epithelial cells, normal (CCR10 and EphA3-poor) and IPF (CCR10 and EphA3-rich) cells were cultured in PEB medium and stimulated with 20 nM preclustered EFNA5-FC (a known ligand for EphA3; refs. 35, 36) in the presence or absence of 10 ng/ml TGF-β1 for 24 hours followed by transcriptomic analysis. EFNA5-FC stimulation significantly increased EphA3 and CCR10 transcripts in IPF relative to normal epithelial cells (Figure 8, E and F). Further, EFNA5-FC promoted TGF-β1–induced expression of basal epithelial cell markers, KRT5 and COL17A1 (Figure 8, G and H), FN1 (Figure 8I), and VIM (Figure 8J) transcripts in IPF relative to normal epithelial cells. These results suggest that EphA3 signaling may be required for the expression of EphA3 and CCR10 on IPF epithelial cells and for TGF-β1–mediated responses by these cells.
Figure 8. EphA3 signaling induces CCR10 and EphA3 transcript expression and promotes TGF-β1–induced VIM and FN1 transcript expression in IPF relative to normal epithelial cells.
(A) Normal and IPF CRC-cultured epithelial cells were stained with fluorescently conjugated anti-EpCAM, -CCR10, and -EphA3 antibodies and analyzed by flow cytometry. Shown are representative dot plots from normal (top) and IPF (bottom) epithelial cultures depicting EpCAM+CCR10+EphA3+ cells. (B) Shown is the percentage of EpCAM+CCR10+EphA3+ cells in normal and IPF epithelial cultures. n = 3–4/group. Data shown are the mean ± SEM. *P ≤ 0.05 via unpaired parametric 2-tailed t test. (C) Magnetic sorting of EphA3+ or EphA3– cells followed by qPCR analysis for various lineage markers. n = 6/group. Data shown are the mean ± SEM. **P ≤ 0.01 via 2-way ANOVA corrected with Bonferroni’s test. (D) RNA was extracted from PEB- or CRC-cultured epithelial cells and qPCR analysis was performed for EphA3. Shown is the average EphA3 expression in CRC-cultured epithelial cells normalized to PEB-cultured cells. n = 3/group. Data shown are the mean ± SEM. *P ≤ 0.05 via unpaired parametric 2-tailed t test with Welch’s correction. (E–J) PEB-cultured normal (n = 2; black bars) and IPF (n = 4; white bars) epithelial cells were stimulated with 20 nM preclustered EFNA5-FC, 10 nM TGF-β, or both for 24 hours. As a control, cells were stimulated with saline (vehicle). After stimulation, RNA was extracted, and qPCR analysis was performed for various transcripts. Depicted is the average expression of CCR10 (E), EphA3 (F), KRT5 (G), COL17A1 (H), FN1 (I), and VIM (J) transcripts in stimulated epithelial cells relative to vehicle-treated cells. Data shown are the mean ± SEM. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 normal versus IPF via 2-way ANOVA corrected with Tukey’s test. IPF, idiopathic pulmonary fibrosis.
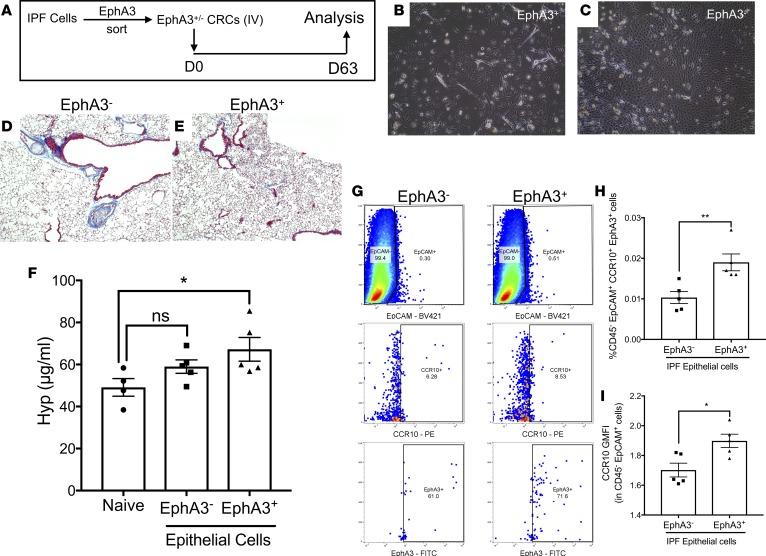
EphA3+ epithelial cells that highly express CCR10 induce more consistent lung remodeling relative to EphA3– epithelial cells in humanized NSG mice.
To confirm the profibrotic potential of EphA3+CCR10+ epithelial cells in vivo, EphA3+ cells were magnetically sorted, and both EphA3+ and EphA3– nonsorted epithelial cells were in vitro expanded in CRC medium (to promote their proliferation) and then intravenously administered into NSG mice. Sixty-three days after administration of sorted and nonsorted cells, mice were sacrificed and their lung tissues were collected for flow cytometric, histological, and biochemical analysis (Figure 9A). Culture-expanded EphA3+ and EphA3– cells showed distinct morphological structures, where the former were hyperplastic and sparsely grew in the cell culture plates (Figure 9B) and the latter grew in distinct colonies with frequent cell-cell contact (Figure 9C). Sixty-three days after cellular administration, EphA3+ cells more consistently induced mild histological interstitial consolidation and remodeling (Figure 9, D and E) and significant increase in hydroxyproline concentration in humanized NSG lungs (Figure 9F) relative to naive, nonhumanized, NSG mice. Consistently, mice receiving EphA3+ epithelial cells had a significantly higher percentage of CD45–EpCAM+CCR10+EphA3+ epithelial cells (Figure 9G and quantified in Figure 9H) and human CD45–EpCAM+ epithelial cells with significantly higher CCR10 GMFI (Figure 9I), relative to mice receiving EphA3– epithelial cells. Collectively, these results suggest that EphA3+ cells, highly expressing CCR10, promote more consistent lung remodeling in humanized NSG mice relative to EphA3– cells and suggest that these epithelial cells may promote fibrotic progression in IPF lungs.
Figure 9. CCR10+EphA3+ epithelial cells induce consistent lung remodeling in NSG mice.
(A) Magnetic sorting of EphA3+ and EphA3– IPF explant cells, followed by expansion of epithelial cells via CRC culture and subsequent i.v. administration into NSG mice. (B and C) Shown are images of CRC-expanded EphA3– (B) and EphA3+ (C) epithelial cells acquired at ×100 magnification. (D and E) Masson’s trichrome staining of NSG lungs 63 days after EphA3– (D) or EphA3+ (E) epithelial cell administration. (F) Depicted is the average hydroxyproline concentration in lungs tissues from naive or humanized NSG mice. Naive: n = 4; IPF CRC: n = 5/group. *P ≤ 0.05 via 1-way ANOVA corrected with Dunnett’s test. (G) Shown are representative dot plots depicting IPF xenografted EpCAM+ epithelial cells (top) staining for CCR10 (middle) and EphA3 (bottom) proteins in IPF EphA3– (left) or EphA3+ (right) epithelial cell–humanized NSG lungs. (H and I) Shown is the percentage of CD45–EpCAM+CCR10+EphA3+ (H) and CCR10 GMFI of CD45–EpCAM+ cells in the lungs of mice humanized with EphA3– or EphA3+ IPF epithelial cells. Data shown are the mean± SEM.; n = 5/group. *P ≤ 0.05; **P ≤ 0.01 via unpaired Mann-Whitney 2-tailed nonparametric test. IPF, idiopathic pulmonary fibrosis; CRC, conditionally reprogrammed cell culture medium; NSG, NOD Cg-PrkdcSCID IL2rgTm1wilSzi.
Discussion
Lung remodeling in IPF is characterized by the expansion of matrix-producing mesenchymal cells and a loss of the alveolar cell–regenerating progenitor, alveolar type II epithelial cells. Despite the loss of alveolar type II epithelial cells, several reports have observed an expansion of airway KRT5+ and SCGB1A1+ cells in the lungs of IPF patients (14–16). However, the role of these epithelial cells in the fibrotic lung progression and/or repair remains elusive. In this manuscript, we demonstrated that there is an expansion of CCR10+EphA3+ epithelial cells in IPF relative to normal explanted lung tissues. Further, using in vitro culture techniques, we demonstrated that epithelial cells cultured from explanted normal and IPF tissues express airway epithelial markers and that conditioned supernatants from IPF epithelial cells promoted IPF lung fibroblast invasion and collagen 1 secretion. Single-cell transcriptomic analysis of FACS-isolated CCR10+ IPF epithelial cells showed that this chemokine receptor is expressed by KRT5+ and SCGB1A1+ epithelial cell populations, both of which showed transcriptomic evidence of mitochondrial dysfunction and proinflammatory and profibrotic pathway enrichment. Consistently, IPF but not normal epithelial cells induced lung remodeling in humanized NSG mice, where EphA3+ epithelial cells that highly expressed CCR10 induced more consistent lung remodeling compared with EphA3– cells that expressed low levels of CCR10. These results describe what we believe is a novel population of epithelial cells in primary human IPF lung tissues that may promote lung remodeling in this fibrotic lung disease.
CCR10 is a C-C chemokine receptor (also known as GPR2), that is expressed on skin- and mucosa-homing T cells (37, 38), IgA-producing plasma cells (39), and antigen-presenting cells (40). While CCR10 has been frequently reported on immune cells, its expression on epithelial cells has been previously reported in murine models of allergic asthma (41) and in pancreatic ductal adenocarcinoma (42). Elevated expression of this chemokine receptor in IPF lungs led us to hypothesize that cells expressing this receptor may promote lung remodeling IPF. However, due to the limited numbers of CCR10+ epithelial cells in normal lung tissues, we were unable to perform studies comparing CCR10+ normal and IPF epithelial cells.
Given the limited recovery of CCR10+ epithelial cells from explanted lung tissues, in vitro culture techniques were developed to expand and characterize these CCR10-enriched epithelial cell cultures. Cells were initially cultured using a modified version the previously described CRC cell culture technique (19, 20, 43), where irradiated feeder cells were replaced with senescent primary lung fibroblast CS. Additionally, cells were cultured in a predefined commercial medium that is optimized to grow airway epithelial cells (i.e., PEB medium). In vitro analysis revealed that senescent conditioned supernatants promoted epithelial cell survival, potentially due to the abundance of hypomethylated mitochondrial DNA (22) and the addition of Y-27632 (which was required for epithelial cell proliferation). This is consistent with other reports observing similar effects with Y27632 on several cell types including airway epithelial cells (44), mesenchymal stem cells (45), astrocytes (46), corneal endothelial cells (47), limbal epithelial cells (48), and keratinocytes (49). In our studies, PEB-cultured cells expressed higher levels of CCR10 and EphA3 transcripts relative to CRC-cultured cells. Cells cultured under CRC conditions were morphologically hyperplastic and less cuboidal, and appeared to proliferate more rapidly, which is consistent with lower expression of the cell cycle regulatory proteins CDKN1A and CDKN1B, relative to PEB-cultured cells. Finally, CRC-cultured cells seemed to revert to cuboidal epithelial cells after 24- to 72-hour culture with PEB medium. It is interesting to note that EphA3 transcript was only detected by qPCR analysis and not in our single-cell RNA sequencing analysis of cultured epithelial cells. We hypothesize that its relatively low expression levels in these cells (as observed via qPCR analysis) may render this transcript below the coverage limit of our single-cell sequencing analysis. Thus, in our studies, CRC medium was utilized to generate enough cells for in vitro and/or in vivo analysis and PEB medium was utilized to promote CCR10 and/or EphA3 expression by these cells.
Physiologically, pulmonary epithelial cells were observed to promote fibroblast quiescence (50, 51); however, under injury conditions, these cells can promote fibroblast proliferation and myofibroblast differentiation (52–55). Our in vitro studies suggest that IPF epithelial cells may promote fibroblast invasion and collagen 1 secretion. This is consistent with one study showing that CCR10 signaling on pancreatic cancer ductal epithelial cells promoted the recruitment and activation of stromal pancreatic stellate cells (42). Further, our in vitro studies suggest that EphA3 signaling promoted CCR10 expression and single-cell transcriptomic analysis of CCR10-expressing IPF epithelial cells showed abundant expression of the CCR10-ligand, CCL28, by these cells (not shown). These results suggest that EphA3 and CCR10 signaling in IPF epithelial cells may cooperate to promote lung remodeling as follows: EphA3 signaling may be required to express or maintain the expression of CCR10 and autocrine CCR10 signaling may induce the secretion of fibroblast recruiting and activation proteins; thus, leading to fibroblast recruitment, activation and subsequent lung remodeling. Unfortunately, we were unable to generate epithelial cells that lacked both transcript and cell-surface expression of CCR10, thus rendering it difficult to utilize a reductionist approach to assess the role of this receptor on the observed epithelial cell phenotype. Future studies are warranted utilizing genetic knockdown and overexpression approaches for CCR10 to better assess the role of this chemokine receptor in airway epithelial cells phenotype and function.
To assess the role of CCR10+ epithelial cells in the promotion of lung remodeling in vivo, a humanized model of this disease was utilized in which immune and structural cells are intravenously administered into NSG mice (17). In this model, CCR10+ human epithelial cells engrafted into the humanized NSG lungs, where the numbers of these cells correlated with hydroxyproline concentration in the remodeled lungs. Similarly, when FACS-isolated CD3–CCR10+ cells were administered into NSG mice, there was a positive correlation between the numbers of CCR10-expressing epithelial cells and hydroxyproline concentration. When magnetically sorted CCR10lo and CCR10hi samples (based on transcript expression) were utilized in this model, only the former group induced lung remodeling after 35 days, and xenografted epithelial cells expressing CCR10 were enriched in the fibrotic lungs of mice receiving CCR10lo cells. This finding is consistent with the reduced expression levels of this chemokine receptor in epithelial relative to immune cells. In separate studies, CCR10-expressing T cells were also observed to promote lung remodeling after 63 days of i.v. administration into NSG mice, but this remodeling response may be dependent on other phenotypic determinants on these cells, including their expression of costimulatory proteins (i.e., CD28 and CTLA4; Habiel et al., in press).
Due to limited recovery of CCR10+ epithelial cells from IPF lung tissues, we were unable to perform NSG studies utilizing FACS-isolated EpCAM+CCR10+ cells. To overcome this limitation, we expanded these cells in vitro prior to administration into NSG mice. Sixty-three days after administration, the number of CCR10+ epithelial cells from IPF, but not normal, lungs correlated with hydroxyproline concentration in fibrotic humanized NSG lung tissues. Consistent with the expression of various mesenchymal markers by single-cell RNA-sequenced CCR10+ epithelial cells, flow cytometric analysis showed that these cells in IPF lungs highly express the receptor tyrosine kinase EphA3, which has been previously reported to be enriched on mesenchymal cells in developing murine lungs (33). Further, a subset of epithelial cells that highly express CCR10 and EphA3 proteins more consistently induced lung remodeling in humanized mice relative to epithelial cells from the same patient expressing low levels of CCR10 transcript and no EphA3 protein. However, given the expression of EphA3 by a minor population of CCR10–epithelial cells from IPF lungs, it is possible that this population of cells may also have contributed to the observed lung remodeling in vivo. Thus, future studies are warranted using xenograft tracking and genetic knockdown/overexpression techniques to better characterize the role of epithelial cells expressing these 2 proteins in lung remodeling.
The contribution of epithelial cells to the pool of fibroblasts and myofibroblasts in fibrotic mouse lungs remains controversial, with some studies showing no contribution (56, 57) and others showing some contribution (58–60) from these cells. KRT5+ cells have been previously observed in the alveolar parenchyma of bleomycin-injured or influenza-infected mouse lungs (12, 13), with one study suggesting that these cells may be distinct from airway basal and club cells (12). In our studies, CCR10+ epithelial cells highly coexpressed a putative mesenchyme-specific receptor tyrosine kinase, EphA3 (33). However, flow cytometric analysis of CCR10+EphA3+ epithelial cells from explanted normal and IPF lung tissues showed that very few epithelial cells expressed another cell surface mesenchymal marker, PDGFRα (8, 34). Further, transcriptomic analysis of mesenchymal marker expression showed that the expression of COL1A1, COL1A2, and COL3A1 transcripts by CCR10+ IPF epithelial cells is minor compared with that observed in fibroblasts (data not shown). This suggests that the contribution of extracellular matrix (ECM) by these cells might be minor compared with the contribution of ECM by professional fibroblasts/myofibroblasts. Thus, profibrotic IPF epithelial cells may indirectly lead to increased matrix deposition in humanized NSG lungs and IPF lungs via the activation of lung fibroblasts.
Collectively, our results have shown that there is significant elevation in the percentage of EpCAM+CCR10+EphA3+ epithelial cells in IPF compared with normal lung tissues. Single-cell transcriptomic analysis showed that these cells were enriched for inflammatory and profibrotic pathways and/or upstream regulators of these pathways. These epithelial cells promoted lung fibroblast invasion and collagen 1 secretion in vitro and lung remodeling in humanized NSG mice in vivo. Collectively, our results suggest that the selective targeting CCR10+ epithelial cells (or profibrotic mediators elaborated by these cells) possibly via EphA3 activity may be beneficial in ameliorating progressive remodeling in IPF.
Methods
Isolation of mixed cells from IPF explants
Normal and IPF lung explants were acquired from consented donors. IPF progression (i.e., rapid versus slow progression) was defined as previously described (61). Fresh explant cells were isolated as previously described (17).
Culturing normal and IPF lung explant–derived airway epithelial cells
Normal and IPF lung epithelial cells were generated by culturing normal or IPF explant suspensions using CRC as previously described (19, 20, 62) with few modifications or in PneumaCult-Ex Plus Medium (STEMCELL Technologies) containing 10 μM Y27632 (STEMCELL Technologies, PEB medium). For CRC culture, normal and IPF explant suspensions were cultured in 50% senescent stromal cell–conditioned supernatants, 50% complete medium (DMEM [Lonza], 15% FBS [Atlanta Biological], 100 IU penicillin and 100 μg/ml streptomycin [Mediatech], 292 μg/ml L-glutamine [Mediatech], and 100 μg/ml Primocin [Invivogen]) and 10 μM Y-27632. Senescence of lung fibroblasts was validated via assessment of morphologic enlargement of the cells, increased β-galactosidase activity and CDKN1A transcript expression. Conditioned supernatants from senescent lung fibroblasts were collected every 3 or 4 days and filtered through a 0.2-μm filter prior to utilization. Explant suspensions were cultured in CRC or PEB media for 2–4 weeks on tissue culture–treated T75 flasks until epithelial colonies were apparent. Contaminating stromal colonies were removed by trypsinizing the cells for 15–20 seconds followed by gentle tapping of the flasks. Once epithelial colonies become relatively dense, cells were detached using Accutase dissociation solution (STEMCELL Technologies) treatment for 10 minutes at 37°C. Cells were passaged 3 or 4 times prior to utilization for flow cytometry, in vitro, or xenograft experiments.
Single-cell RNA sequencing and bioinformatic analysis
IPF explant suspensions were cultured in CRC medium until epithelial colonies were apparent, and cells were then transferred into PEB medium overnight and dissociated using Accutase dissociation solution as described above (Culturing normal and IPF lung explant derived airway epithelial cells section). Cells were then stained with BV421 anti-EpCAM (clone 9C4, BioLegend) and PE anti-CCR10 antibodies (clone 6588-5, BioLegend). EpCAM+CCR10+ and EpCAM+CCR10– epithelial cells were then FACS isolated using a FACSAria III cell sorter (BD Biosciences). Sorted epithelial cells were spun down, counted, and resuspended and single-cell sequencing was performed as recommended by the manufacturer (10× Genomics). All sequencing files have been uploaded onto the NCBI’s Gene Expression Omnibus (GEO GSE115982). Detailed methods pertaining to the single-cell data analysis are provided in the online supplement.
In vitro assessment of EphA3 signaling and epithelial-stromal cross-talk
Normal and IPF epithelial cells were cultured at a concentration of 0.4 × 106 cells/well in a 6-well plate in PEB medium. EFNA5-FC (R&D Systems) was preclustered by incubating it at an EFNA5-FC/anti–His-tag antibody (R&D Systems) ratio of 10:1 in Dulbecco’s PBS (DPBS) for approximately 20 minutes at room temperature. Epithelial cells were then stimulated with 20 nM preclustered EFNA5-FC, 10 ng/ml TGF-β1 (R&D Systems), or a combination of the stimuli. Cells were then incubated for 24 hours at 37°C and 10% CO2. As a control, similar stimuli were added to a cell-free 6-well plate and incubated for 24 hours. After incubation, conditioned supernatants were collected and frozen at –20°C until utilization with fibroblast studies and 1 ml of TRIzol (Thermo Fisher Scientific) was added to the stimulated epithelial cells. RNA was extracted from the epithelial cells and utilized for qPCR analysis.
Fibroblast invasion analysis
Lung fibroblasts were generated from explanted lung tissues as previously described (63). Conditioned supernatants from normal or IPF epithelial cells were added to lung fibroblasts and fibroblast invasion was assessed as previously described (63).
Mice and in vivo studies
Six- to 8-week-old, female, pathogen-free NOD Cg-PrkdcSCID IL2rgTm1wilSzi (NSG) or NOD.Cg-PrkdcSCID IL2rgtm1Wil Tg (CAG-EGFP) 10sb/SzJ (NSG-GFP) mice were purchased from Jackson Laboratories and housed in Cedars-Sinai Medical Center’s high isolation animal rooms. NSG mice were allowed a minimum of 1 week to acclimate in the facility and then each mouse received nothing (i.e., nonhumanized); 1 × 106 IPF explant cells; 1.5 × 105 CD3–CCR10+ cells; 2.5 × 105 CCR10lo or CCR10hi cells; 5 × 105 cultured normal or IPF epithelial cells or 8 × 104 cultured CCR10hiEphA3+ or CCR10loEphA3– epithelial cells by intravenous injection (i.e., humanized). In vivo studies with explanted lung cells utilized bio-banked cells from liquid N2 storage that were rapidly thawed and washed in serum-free medium prior to injection into NSG mice. All NSG groups were monitored daily and mice were sacrificed if there was evidence of morbidity such as weight loss of more than 20%, loss of fur, paralysis and/or lack of responsiveness when handled. At the indicated times after human cell injection, bronchoalveolar lavage (BAL) fluid and serum were collected for protein analysis, the superior and middle lobes were collected for biochemical hydroxyproline quantification, the inferior lobe and spleens for flow cytometric analysis, post-caval lobe for qPCR analysis, and the left lung for histological analysis from each NSG mouse. For studies where cells were magnetically sorted prior to administration into NSG mice, lung explant cells were stained with PE-conjugated anti-CCR10 antibodies (clone 6588-5, BioLegend) and then magnetically sorted using anti-PE microbeads (Miltenyi Biotec or STEMCELL Technologies) as recommended by the manufacturer. Enrichment for CCR10+ cells was confirmed by qPCR analysis for CCR10 transcript in sorted versus nonsorted normal and IPF cells. For humanized NSG experiments, 0.25 million sorted cells/mouse were i.v. administered into NSG mice. Mice were monitored daily for any clinical signs of distress and/or morbidity. Thirty-five days after cellular administration, the mice were sacrificed, and their lung tissues were collected and analyzed as indicated in the mouse studies methods section.
Hydroxyproline assay
Lung tissue was hydrolyzed and total lung hydroxyproline was analyzed as previously described (64). To compare changes in hydroxyproline concentration, humanized lung samples were assayed together with hydrolyzed lung samples from naive mice housed in the same facility and treated similarly (17).
Histological analysis
Lung tissue was fixed in 10% neutral buffered formalin (NBF) solution for 24 hours and subsequently transferred into tissue cassettes and kept in a 70% ethanol solution for approximately 24 hours. Lungs were then processed using routine histological techniques and stained using Masson’s trichrome. A Zeiss Axio Observer Z1 microscope and the Zeiss Zen 2012 v.1.1.2.0 software were used to obtain representative images.
Flow cytometry
Mouse lung cell suspensions.
Murine lung cellular suspensions were generated using a mouse lung dissociation kit, C-tubes, and a GentleMacs dissociator (Miltenyi Biotec). Cell suspensions were centrifuged and resuspended in 1 ml of RBC lysis buffer (BioLegend), incubated at room temperature for 1–2 minutes and then equilibrated by adding 25 ml of DPBS to each tube. Cells were centrifuged and resuspended in flow cytometric wash/staining buffer (DPBS + 2% FBS) in the presence of human and mouse Fc receptor–blocking antibodies (BioLegend). Cells were then stained with anti–human CD45 (clone HI30, BioLegend), -EpCAM (clone 9C4, BioLegend) and -CCR10 (clone 6588-5, BioLegend) antibodies for 20 minutes at 4°C. Unstained, isotype controls, fluorescence-minus-one (FMO) controls, and staining in nonhumanized murine lung and spleen suspensions were utilized to gate out any nonspecific antibody binding and background fluorescence (Supplemental Figures 8 and 9). Flow cytometric data were acquired within 1 week of staining using a MACSQuant 10 flow cytometer (Miltenyi Biotec) and data were analyzed using FlowJo software v.10.2 (Tree Star). Due to the presence of a population of GFP– cells in naive, nonhumanized NSG-GFP mice, all of our comparisons were performed relative to nonhumanized NSG-GFP mice. The resulting gates from nonhumanized NSG-GFP mice were used to identify human EPCAM- and CCR10-stained cells.
Human lung cell suspensions.
Human lung cells were added to flow cytometry wash/staining buffer and blocked with anti–human Fc receptor antibodies (BioLegend) for 15 minutes at 4°C. After blocking, cells were stained with PE-conjugated anti–human CCR10 (clone 6588-5, BioLegend, 341504), BV421- or APC-conjugated anti–human EpCAM (clone 9C4, BioLegend, 324220 and 324208, respectively), PE/Cy7- or BV421-conjugated anti–human CD45 (clone HI30, BioLegend, 304016 and 304032, respectively), PE/Cy7-conjugated anti-PDGFRα (clone 16A1, BioLegend, 323508) or biotin-conjugated anti–human EphA3 (clone IIIA4, supplied by Humanigen, Inc.) and FITC-conjugated streptavidin (Thermo Fisher Scientific) for 20 minutes at 4°C. Cells were then washed twice with flow cytometry wash/staining buffer and fixed in 5% NBF. Flow cytometric data were acquired using a MACSQuant 10 flow cytometer and data were analyzed using FlowJo software v.10.2. Cells were gated using isotype and/or FMO controls as shown in Supplemental Figure 10.
Cultured epithelial cells.
CRC- or PEB-media-cultured epithelial cells were dissociated using Accutase cell dissociation solution. Cells were washed, resuspended in flow cytometric wash/staining buffer and blocked with anti–human Fc receptor antibodies (BioLegend) for 15 minutes at 4°C. After blocking, cells were stained with PE-conjugated anti–human CCR10 (clone 6588-5, BioLegend, 341504), BV421- or APC-conjugated anti–human EpCAM (clone 9C4, BioLegend, 324220 and 324208, respectively) or biotin-conjugated anti–human EphA3 mAb (clone IIIA4, supplied by Humanigen, Inc.) and FITC-conjugated streptavidin (Thermo Fisher Scientific) for 20 minutes at 4°C. Cells were then washed twice with flow cytometry wash/staining buffer and fixed in 5% NBF. Flow cytometric data were acquired using a MACSQuant 10 flow cytometer and data were analyzed using FlowJo software v.10.2.
Magnetic epithelial cell sorting for CCR10
Lung epithelial cells were dissociated using Accutase cell dissociation solution and spun down as described above (Cell culture and culture conditions). Cells were resuspended at a concentration of 5 × 106 cells/ml and stained with biotinylated anti-EphA3 antibodies (clone IIIA4, supplied by Humanigen, Inc.). After staining, cells were washed and then magnetically sorted using streptavidin magnetic beads (Miltenyi Biotec) as recommended by the manufacturer.
qPCR analysis
Cells were lysed in TRIzol reagent. RNA was extracted as recommended by the manufacturer and 1 μg of RNA was reverse transcribed into cDNA using Superscript II reverse transcriptase (Life Technologies) as previously described (61). Complementary DNA (cDNA) was subsequently loaded into a Taqman plate (Thermo Fisher Scientific) and gene expression analyses were performed using predesigned primers and probes for human CCR10, KRT5, MUC5B, SCGB1A1, SFTPC, EphA3, CDKN1A, CDKN1B, IL1A, IL1B, IL6, TGFB1, TNF, COL17A1, FN1, VIM, and RNA18S5 (Thermo Fisher Scientific). All Taqman analyses were performed using an Applied Biosystems Viia 7 instrument (Thermo Fisher Scientific). The results were then exported, normalized to RNA18S5 expression (Thermo Fisher Scientific), and fold-change values were calculated using DataAssist software (Thermo Fisher Scientific).
Statistics
Two-tailed parametric t test, 1-tailed Mann-Whitney nonparametric test, 2-tailed Mann-Whitney nonparametric test, 1-way ANOVA or 2-way ANOVA statistical analyses (as indicated in the figure legends) were performed using GraphPad Prism software version 7. Multi-sample comparison was corrected using Dunnett’s test, when comparing to a normal or vehicle control, or Tukey’s test when comparing all samples to each other. P ≤ 0.05 was considered significant.
Study approval
The Institutional Review Board at Cedars-Sinai Medical Center approved all experiments with primary human tissue. Informed consent was obtained from all patients prior to inclusion in the studies described herein. Cedars-Sinai Medical Center Department of Comparative Medicine and the University of Michigan Unit for Laboratory Animal Medicine approved all mouse studies described herein. All studies were performed in accordance with the relevant guidelines and regulations.
Author contributions
DMH conceived experiments, performed experiments, analyzed results, and wrote and edited the manuscript. MSE performed experiments and edited the manuscript. ALC and ICJ performed experiments. BS edited the manuscript and helped analyze results. CMH conceived experiments and wrote and edited the manuscript.
Supplementary Material
Acknowledgments
These studies were supported by funds and reagents from Humanigen Inc., Cedars-Sinai Medical Center, and the NIH (R01HL123899). D.M. Habiel was supported by Cedars-Sinai Medical Center. The authors would like to also acknowledge Kathy McClinchey and McClinchey Histology Lab Inc. for their histological services, and Yizhou Wang, Chintda Santiskulvong, and Jie Tang from the Cedars-Sinai genomics core. Additionally, we would like to acknowledge the biobank and flow cytometric cores and members of Barry Stripp’s laboratory for their help in acquiring the patient and donor tissues.
Version 1. 08/23/2018
Electronic publication
Funding Statement
David Habiel & Cory Hogaboam were supported by Cedars-Sinai Medical Center
This work was supported by Cory Hogaboam’s NHLBI grant number R01HL123899.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(16):e122211. https://doi.org/10.1172/jci.insight.122211.
Contributor Information
Ana Lucia Coelho, Email: AnaLucia.Coelho@cshs.org.
Cory M. Hogaboam, Email: Cory.Hogaboam@cshs.org.
References
- 1.Esposito DB, et al. Idiopathic pulmonary fibrosis in united states automated claims. Incidence, prevalence, and algorithm validation. Am J Respir Crit Care Med. 2015;192(10):1200–1207. doi: 10.1164/rccm.201504-0818OC. [DOI] [PubMed] [Google Scholar]
- 2.Raghu G, Chen SY, Hou Q, Yeh WS, Collard HR. Incidence and prevalence of idiopathic pulmonary fibrosis in US adults 18-64 years old. Eur Respir J. 2016;48(1):179–186. doi: 10.1183/13993003.01653-2015. [DOI] [PubMed] [Google Scholar]
- 3.Raimundo K, Chang E, Broder MS, Alexander K, Zazzali J, Swigris JJ. Clinical and economic burden of idiopathic pulmonary fibrosis: a retrospective cohort study. BMC Pulm Med. 2016;16:2. doi: 10.1186/s12890-015-0165-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King TE, et al. A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N Engl J Med. 2014;370(22):2083–2092. doi: 10.1056/NEJMoa1402582. [DOI] [PubMed] [Google Scholar]
- 5.Noble PW, et al. Pirfenidone in patients with idiopathic pulmonary fibrosis (CAPACITY): two randomised trials. Lancet. 2011;377(9779):1760–1769. doi: 10.1016/S0140-6736(11)60405-4. [DOI] [PubMed] [Google Scholar]
- 6.Mazzei ME, Richeldi L, Collard HR. Nintedanib in the treatment of idiopathic pulmonary fibrosis. Ther Adv Respir Dis. 2015;9(3):121–129. doi: 10.1177/1753465815579365. [DOI] [PubMed] [Google Scholar]
- 7.Richeldi L, et al. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N Engl J Med. 2014;370(22):2071–2082. doi: 10.1056/NEJMoa1402584. [DOI] [PubMed] [Google Scholar]
- 8.Ntokou A, et al. Characterization of the platelet-derived growth factor receptor-α-positive cell lineage during murine late lung development. Am J Physiol Lung Cell Mol Physiol. 2015;309(9):L942–L958. doi: 10.1152/ajplung.00272.2014. [DOI] [PubMed] [Google Scholar]
- 9.Green J, Endale M, Auer H, Perl AK. Diversity of interstitial lung fibroblasts is regulated by platelet-derived growth factor receptor α kinase activity. Am J Respir Cell Mol Biol. 2016;54(4):532–545. doi: 10.1165/rcmb.2015-0095OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hung C, et al. Role of lung pericytes and resident fibroblasts in the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med. 2013;188(7):820–830. doi: 10.1164/rccm.201212-2297OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Agha E, et al. Two-way conversion between lipogenic and myogenic fibroblastic phenotypes marks the progression and resolution of lung fibrosis. Cell Stem Cell. 2017;20(4):571. doi: 10.1016/j.stem.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 12.Ray S, et al. Rare SOX2+ airway progenitor cells generate KRT5+ cells that repopulate damaged alveolar parenchyma following influenza virus infection. Stem Cell Reports. 2016;7(5):817–825. doi: 10.1016/j.stemcr.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaughan AE, et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. 2015;517(7536):621–625. doi: 10.1038/nature14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight. 2016;1(20):e90558. doi: 10.1172/jci.insight.90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smirnova NF, Schamberger AC, Nayakanti S, Hatz R, Behr J, Eickelberg O. Detection and quantification of epithelial progenitor cell populations in human healthy and IPF lungs. Respir Res. 2016;17(1):83. doi: 10.1186/s12931-016-0404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buendía-Roldán I, et al. Increased expression of CC16 in patients with idiopathic pulmonary fibrosis. PLoS One. 2016;11(12):e0168552. doi: 10.1371/journal.pone.0168552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Habiel DM, Espindola MS, Coelho AL, Hogaboam CM. Modeling idiopathic pulmonary fibrosis in humanized severe combined immunodeficient mice. Am J Pathol. 2018;188(4):891–903. doi: 10.1016/j.ajpath.2017.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Habiel D, et al. Characterization of profibrotic Ccr10+Epha3+ cells in IPF lungs. Am J Respir Crit Care Med. 2017;195:A7072 [Google Scholar]
- 19.Saenz FR, et al. Conditionally reprogrammed normal and transformed mouse mammary epithelial cells display a progenitor-cell-like phenotype. PLoS One. 2014;9(5):e97666. doi: 10.1371/journal.pone.0097666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suprynowicz FA, et al. Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc Natl Acad Sci USA. 2012;109(49):20035–20040. doi: 10.1073/pnas.1213241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palechor-Ceron N, et al. Radiation induces diffusible feeder cell factor(s) that cooperate with ROCK inhibitor to conditionally reprogram and immortalize epithelial cells. Am J Pathol. 2013;183(6):1862–1870. doi: 10.1016/j.ajpath.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryu C, et al. Extracellular mitochondrial DNA is generated by fibroblasts and predicts death in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2017;196(12):1571–1581. doi: 10.1164/rccm.201612-2480OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Álvarez D, et al. IPF lung fibroblasts have a senescent phenotype. Am J Physiol Lung Cell Mol Physiol. 2017;313(6):L1164–L1173. doi: 10.1152/ajplung.00220.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitasato Y, et al. Enhanced expression of interleukin-18 and its receptor in idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol. 2004;31(6):619–625. doi: 10.1165/rcmb.2003-0306OC. [DOI] [PubMed] [Google Scholar]
- 25.Xie T, et al. Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep. 2018;22(13):3625–3640. doi: 10.1016/j.celrep.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jakubzick C, Kunkel SL, Puri RK, Hogaboam CM. Therapeutic targeting of IL-4- and IL-13-responsive cells in pulmonary fibrosis. Immunol Res. 2004;30(3):339–349. doi: 10.1385/IR:30:3:339. [DOI] [PubMed] [Google Scholar]
- 27.Jakubzick C, et al. Human pulmonary fibroblasts exhibit altered interleukin-4 and interleukin-13 receptor subunit expression in idiopathic interstitial pneumonia. Am J Pathol. 2004;164(6):1989–2001. doi: 10.1016/S0002-9440(10)63759-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jakubzick C, et al. Therapeutic attenuation of pulmonary fibrosis via targeting of IL-4- and IL-13-responsive cells. J Immunol. 2003;171(5):2684–2693. doi: 10.4049/jimmunol.171.5.2684. [DOI] [PubMed] [Google Scholar]
- 29.Murray LA, et al. Targeting interleukin-13 with tralokinumab attenuates lung fibrosis and epithelial damage in a humanized SCID idiopathic pulmonary fibrosis model. Am J Respir Cell Mol Biol. 2014;50(5):985–994. doi: 10.1165/rcmb.2013-0342OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yokoba M, et al. Corrigendum to “Aminophylline increases respiratory muscle activity during hypercapnia in humans” [Pulm. Pharmacol. Ther. (2015) 96-101] Pulm Pharmacol Ther. 2016;41:103. doi: 10.1016/j.pupt.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 31.Vasakova M, et al. IL-4 polymorphisms, HRCT score and lung tissue markers in idiopathic pulmonary fibrosis. Hum Immunol. 2013;74(10):1346–1351. doi: 10.1016/j.humimm.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 32.Park SW, et al. Interleukin-13 and its receptors in idiopathic interstitial pneumonia: clinical implications for lung function. J Korean Med Sci. 2009;24(4):614–620. doi: 10.3346/jkms.2009.24.4.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lahtela J, et al. The putative tumor suppressor gene EphA3 fails to demonstrate a crucial role in murine lung tumorigenesis or morphogenesis. Dis Model Mech. 2015;8(4):393–401. doi: 10.1242/dmm.019257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zepp JA, et al. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell. 2017;170(6):1134–1148.e10. doi: 10.1016/j.cell.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lackmann M, et al. Distinct subdomains of the EphA3 receptor mediate ligand binding and receptor dimerization. J Biol Chem. 1998;273(32):20228–20237. doi: 10.1074/jbc.273.32.20228. [DOI] [PubMed] [Google Scholar]
- 36.Forse GJ, et al. Distinctive structure of the EphA3/Ephrin-A5 complex reveals a dual mode of Eph receptor interaction for Ephrin-A5. PLoS One. 2015;10(5):e0127081. doi: 10.1371/journal.pone.0127081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gosling J, et al. Cutting edge: identification of a novel chemokine receptor that binds dendritic cell- and T cell-active chemokines including ELC, SLC, and TECK. J Immunol. 2000;164(6):2851–2856. doi: 10.4049/jimmunol.164.6.2851. [DOI] [PubMed] [Google Scholar]
- 38.Hudak S, et al. Immune surveillance and effector functions of CCR10(+) skin homing T cells. J Immunol. 2002;169(3):1189–1196. doi: 10.4049/jimmunol.169.3.1189. [DOI] [PubMed] [Google Scholar]
- 39.Kunkel EJ, et al. CCR10 expression is a common feature of circulating and mucosal epithelial tissue IgA Ab-secreting cells. J Clin Invest. 2003;111(7):1001–1010. doi: 10.1172/JCI17244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sisirak V, et al. CCR6/CCR10-mediated plasmacytoid dendritic cell recruitment to inflamed epithelia after instruction in lymphoid tissues. Blood. 2011;118(19):5130–5140. doi: 10.1182/blood-2010-07-295626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.English K, Brady C, Corcoran P, Cassidy JP, Mahon BP. Inflammation of the respiratory tract is associated with CCL28 and CCR10 expression in a murine model of allergic asthma. Immunol Lett. 2006;103(2):92–100. doi: 10.1016/j.imlet.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 42.Roy I, et al. Cancer cell chemokines direct chemotaxis of activated stellate cells in pancreatic ductal adenocarcinoma. Lab Invest. 2017;97(3):302–317. doi: 10.1038/labinvest.2016.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gentzsch M, et al. Pharmacological rescue of conditionally reprogrammed cystic fibrosis bronchial epithelial cells. Am J Respir Cell Mol Biol. 2017;56(5):568–574. doi: 10.1165/rcmb.2016-0276MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horani A, Nath A, Wasserman MG, Huang T, Brody SL. Rho-associated protein kinase inhibition enhances airway epithelial Basal-cell proliferation and lentivirus transduction. Am J Respir Cell Mol Biol. 2013;49(3):341–347. doi: 10.1165/rcmb.2013-0046TE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakamura K, Yoshimura A, Kaneko T, Sato K, Hara Y. ROCK inhibitor Y-27632 maintains the proliferation of confluent human mesenchymal stem cells. J Periodont Res. 2014;49(3):363–370. doi: 10.1111/jre.12114. [DOI] [PubMed] [Google Scholar]
- 46.Yu Z, Liu M, Fu P, Xie M, Wang W, Luo X. ROCK inhibition with Y27632 promotes the proliferation and cell cycle progression of cultured astrocyte from spinal cord. Neurochem Int. 2012;61(7):1114–1120. doi: 10.1016/j.neuint.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 47.Peh GS, et al. The effects of Rho-associated kinase inhibitor Y-27632 on primary human corneal endothelial cells propagated using a dual media approach. Sci Rep. 2015;5:9167. doi: 10.1038/srep09167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun CC, Chiu HT, Lin YF, Lee KY, Pang JH. Y-27632, a ROCK inhibitor, promoted limbal epithelial cell proliferation and corneal wound healing. PLoS One. 2015;10(12):e0144571. doi: 10.1371/journal.pone.0144571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chapman S, Liu X, Meyers C, Schlegel R, McBride AA. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J Clin Invest. 2010;120(7):2619–2626. doi: 10.1172/JCI42297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Epa AP, et al. Normal human lung epithelial cells inhibit transforming growth factor-β induced myofibroblast differentiation via prostaglandin E2. PLoS One. 2015;10(8):e0135266. doi: 10.1371/journal.pone.0135266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kohyama T, et al. Prostaglandin E(2) inhibits fibroblast chemotaxis. Am J Physiol Lung Cell Mol Physiol. 2001;281(5):L1257–L1263. doi: 10.1152/ajplung.2001.281.5.L1257. [DOI] [PubMed] [Google Scholar]
- 52.Mio T, et al. Human bronchial epithelial cells modulate collagen gel contraction by fibroblasts. Am J Physiol. 1998;274(1 Pt 1):L119–L126. doi: 10.1152/ajplung.1998.274.1.L119. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, et al. BRS-3 activation transforms the effect of human bronchial epithelial cells from PGE2 mediated inhibition to TGF-beta1 dependent promotion on proliferation and collagen synthesis of lung fibroblasts. Cell Biol Int. 2007;31(12):1495–1500. doi: 10.1016/j.cellbi.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 54.Zhang S, Smartt H, Holgate ST, Roche WR. Growth factors secreted by bronchial epithelial cells control myofibroblast proliferation: an in vitro co-culture model of airway remodeling in asthma. Lab Invest. 1999;79(4):395–405. [PubMed] [Google Scholar]
- 55.Dosanjh A, Morris RE, Wan B. Bronchial epithelial cell-derived cytokine IL-10 and lung fibroblast proliferation. Transplant Proc. 2001;33(1-2):352–354. doi: 10.1016/S0041-1345(00)02044-3. [DOI] [PubMed] [Google Scholar]
- 56.Rock JR, et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci USA. 2011;108(52):E1475–E1483. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamada M, et al. Dual-immunohistochemistry provides little evidence for epithelial-mesenchymal transition in pulmonary fibrosis. Histochem Cell Biol. 2008;129(4):453–462. doi: 10.1007/s00418-008-0388-9. [DOI] [PubMed] [Google Scholar]
- 58.Tanjore H, et al. Contribution of epithelial-derived fibroblasts to bleomycin-induced lung fibrosis. Am J Respir Crit Care Med. 2009;180(7):657–665. doi: 10.1164/rccm.200903-0322OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim KK, et al. Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc Natl Acad Sci USA. 2006;103(35):13180–13185. doi: 10.1073/pnas.0605669103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu Z, et al. Detection of epithelial to mesenchymal transition in airways of a bleomycin induced pulmonary fibrosis model derived from an alpha-smooth muscle actin-Cre transgenic mouse. Respir Res. 2007;8:1. doi: 10.1186/1465-9921-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trujillo G, et al. TLR9 differentiates rapidly from slowly progressing forms of idiopathic pulmonary fibrosis. Sci Transl Med. 2010;2(57):57ra82. doi: 10.1126/scitranslmed.3001510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao B, et al. Non-malignant respiratory epithelial cells preferentially proliferate from resected non-small cell lung cancer specimens cultured under conditionally reprogrammed conditions. Oncotarget. 2017;8(7):11114–11126. doi: 10.18632/oncotarget.14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Espindola MS, et al. Targeting of TAM receptors ameliorates fibrotic mechanisms in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2018;197(11):1443–1456. doi: 10.1164/rccm.201707-1519OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Habiel DM, Espindola MS, Coelho AL, Hogaboam CM. Modeling idiopathic pulmonary fibrosis in humanized severe combined immunodeficient mice. Am J Pathol. 2018;188(4):891–903. doi: 10.1016/j.ajpath.2017.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.