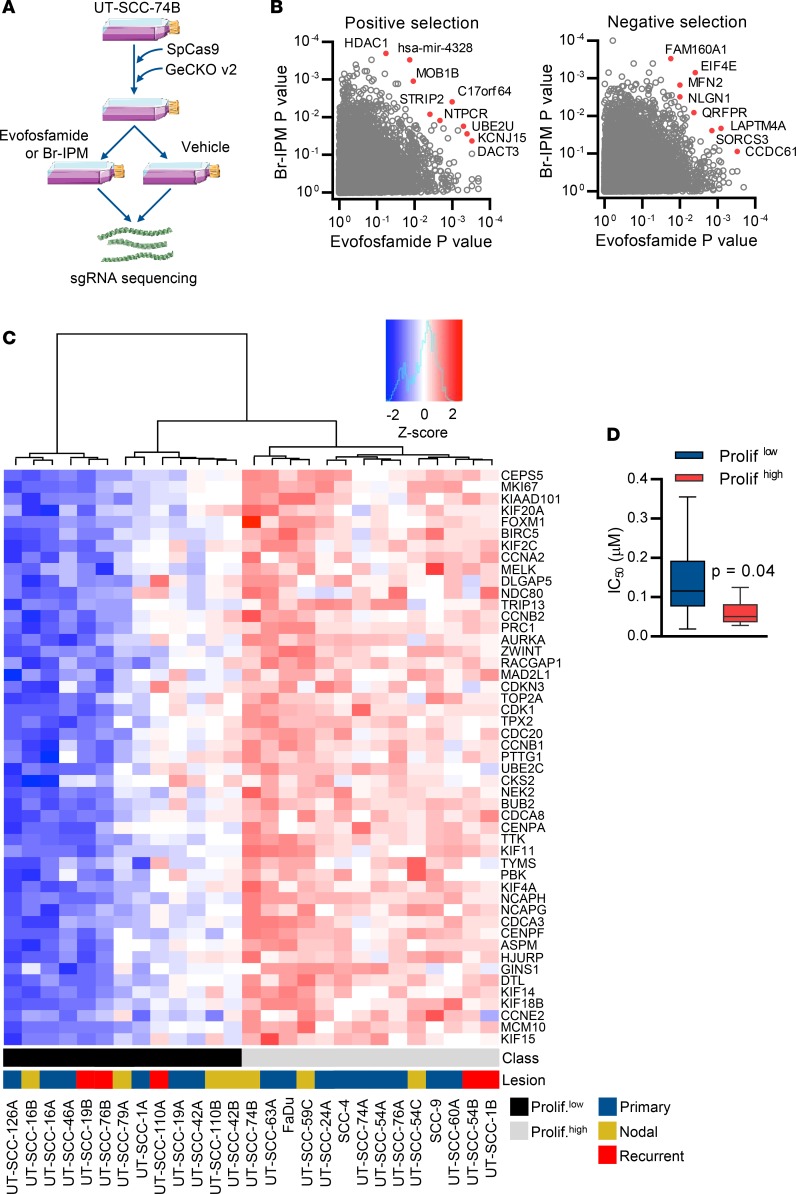
Figure 4. A proliferation expression signature correlates with evofosfamide sensitivity in head and neck squamous cell carcinoma (HNSCC) cell lines.
(A) The workflow for functional genomic screens for modifiers of evofosfamide or Br-IPM sensitivity. SpCas9, Streptococcus pyogenes Cas9. (B) Enrichment (positive selection) or depletion (negative selection) of single guide RNA (sgRNA) in evofosfamide- and Br-IPM-treated UT-SCC-74B cells (2 replicates per condition). The screen was deconvoluted using the RIGER method (83) with weighted-sum aggregation to output gene-level P values that were adjusted using the Benjamini-Hochberg method. (C) Unsupervised hierarchical clustering (ward.D method with Euclidean distance) of HNSCC cell lines according to their expression (measured by RNAseq) of a published tumor proliferation metagene (57). Of 61 genes in the proliferation cluster, 49 were expressed in HNSCC cells. The resulting proliferation metagene class assignments and whether cell lines were derived from primary, nodal, or recurrent lesions are indicated. (D) Differential in vitro sensitivity to evofosfamide per antiproliferative IC50 assay in HNSCC cell lines separated by proliferation metagene status, where the grouping of cell lines as proliferationhi or proliferationlo was accomplished by assignment of cell lines to binary clusters defined by unsupervised hierarchical clustering according to expression values of the metagene as shown in C. Box plots show the mean and interquartile range, whereas whiskers show the maximum and minimum IC50 values for metagene-high (n = 13) and metagene-low (n = 8) cell lines (≥3 independent experiments). Statistical significance was assessed by Mann–Whitney test.