Figure 2.
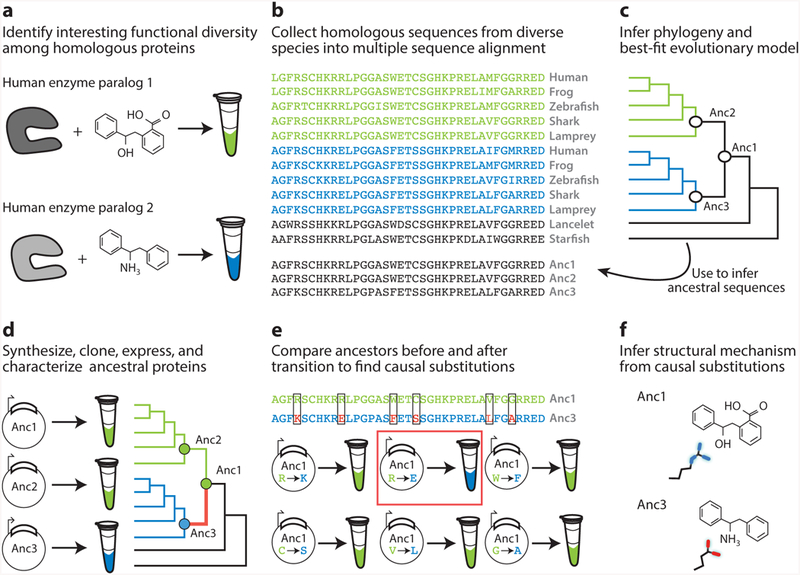
Workflow for vertical analysis of the genetic and structural causes of functional differences between related proteins, shown for a hypothetical family of enzymes. (a) Two paralogous enzymes catalyze similar reactions on different substrates, yielding different products (colors). (b) Sequences of both paralogs ( green and blue) are collected and aligned from many species, including outgroups (black). (c) The alignment is used to computationally infer the best-fit evolutionary model and a phylogeny. Ancestral sequences are inferred by maximum likelihood at nodes representing the last common ancestor of each paralog group (Anc2, Anc3) and at the gene duplication ancestral to both groups. (d ) DNA sequences coding for ancestral proteins are synthesized and cloned; ancestral proteins are expressed and their functions experimentally characterized. This allows the branch on which a new function evolved (red ) to be identified. (e) The substitutions that conferred the derived (blue) function must be among the differences between Anc1 and Anc3 (boxed sites). To identify causal substitutions, amino acid states from Anc3 (red states in blue sequence) are introduced into Anc1 and the resulting proteins tested experimentally (bottom). In the example, an arginine to glutamate substitution (red box) recapitulates the switch in specificity. ( f ) Structures or homology models of ancestral proteins are determined to infer the mechanism by which causal substitutions conferred the new function. In this case, the derived glutamate of Anc3 satisfied the hydrogen bonding potential of the amine group unique to the derived ligand.
