Figure 4.
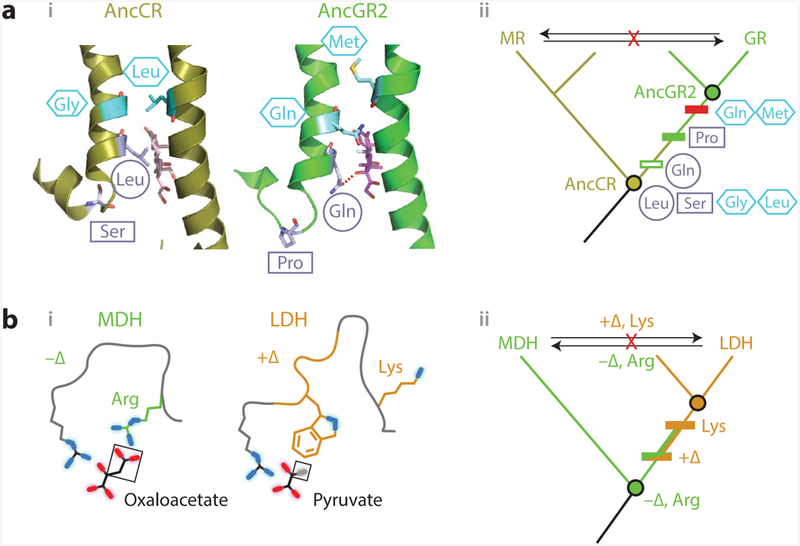
Vertical analysis has illuminated mechanisms of epistasis and functional change in protein evolution.(a) Evolution of hormone specificity in glucocorticoid and mineralocorticoid receptors. (i, left) Position of key helices in the crystal structure of AncCR, the ancestor of all MRs and GRs (olive; 2Q1H) with aldosterone bound (sticks); (right) position of helices in AncGR2, the ancestor of cortisol-specific GRs ( green; 3GN8), with cortisol bound (59). Sites that change specificity are shown as light blue sticks. The Ser–Pro substitution (rectangle) repositions one helix, allowing the Leu–Gln substitution (circle) to form a cortisol-specific hydrogen bond (dashed red line). Restrictive substitutions (hexagons) introduce residues into AncGR2 that would clash in the ancestral helix conformation (13). (ii ) Phylogeny showing the inferred historical order of the substitutions between AncCR and AncGR. Horizontally swapping key residues between paralogs (arrows) yields nonfunctional proteins. (b) Evolution of substrate specificity in apicomplexan malate and lactate dehydrogenases (11). (i ) Active-site geometry of extant DHs, with the key side chain and loop insertion (+Δ) highlighted in green and orange. Substrates (black lines, with oxygen atoms in red and methyl group in gray) are labeled; functional groups unique to each substrate are boxed. (ii ) Phylogeny and ancestral reconstruction showed that LDH function (orange) evolved from an MDH-like ancestor (green). Introducing the derived loop and Lysine residue into the deepest ancestor confers pyruvate specificity. Horizontal swaps of these features (arrows) failed to confer on either protein its paralog’s functional specificity. Abbreviations: AncCR, ancestral corticoid receptor; AncGR, ancestral glucocorticoid receptor; Arg, arginine; DH, dehydrogenase; Gln, glutamine; Gly, glycine; GR, glucocorticoid receptors; LDH, lactate dehydrogenase; Leu, leucine; Lys, lysine; MDH, malate dehydrogenase; Met, methionine; MR, mineralocorticoid; Pro, proline; Ser, serine.
