Figure 5.
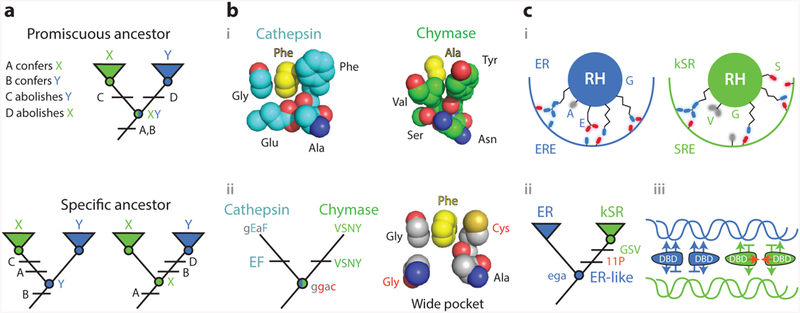
Knowledge of ancestral states clarifies structure-function mechanisms. (a) Simplified example of the implications of vertical analysis. Homologs with distinct functions X and Y can be generated by partitioning functions from a multifunctional ancestral protein (top) or by a discrete change in function from a specific ancestor (bottom). Different trajectories imply different effects of key sequence differences (A–D). (b) Mechanism of evolution of serine protease specificity. (i ) Specialized tight binding pockets of the extant serine proteases cathepsin (1CGH) and chymase (2RDL). (ii ) Their reconstructed last common ancestor had both activities and a wide binding pocket (79). Lower- and upper-case letters show ancestral and derived amino acid states for key residues, using the single-letter code. Ancestral states that confer the promiscuous wide pocket are highlighted in red. (c) Evolution of DNA specificity in steroid hormone receptors. Estrogen and ketosteroid receptors bind different DNA sequences (ERE and SRE). (i ) Schematic of the receptors’ recognition helices bound to the DNA major groove. Residues at variable sites are labeled. kSRs ( green) make fewer specific interactions than ERs (blue). (ii ) Vertical analysis showed that ERs and kSRs evolved from an ER-like ancestor (57). Specificity-switching substitutions (ancestral ega to derived GSV in single-letter code) and permissive substitutions (11P) are labeled.(iii ) Interactions that characterize ER/ERE (blue) and kSR/SRE ( green) complexes are shown, with favorable interactions as arrows and exclusionary interactions as horizontal lines. Permissive substitutions enhanced dimer formation and cooperativity of binding (red arrows) in kSRs. Abbreviations: Ala, alanine; Asn, asparagine; Cys, cysteine; DBD, DNA-binding domain; ER, estrogen receptors; ERE, estrogen response element; Glu, glutamic acid; Gly, glycine; kSR, ketosteroid receptors; Phe, phenylalanine; RH, recognition helice; Ser, seranine; SRE, steroid response element; Tyr, tyrosine; Val, valine.
