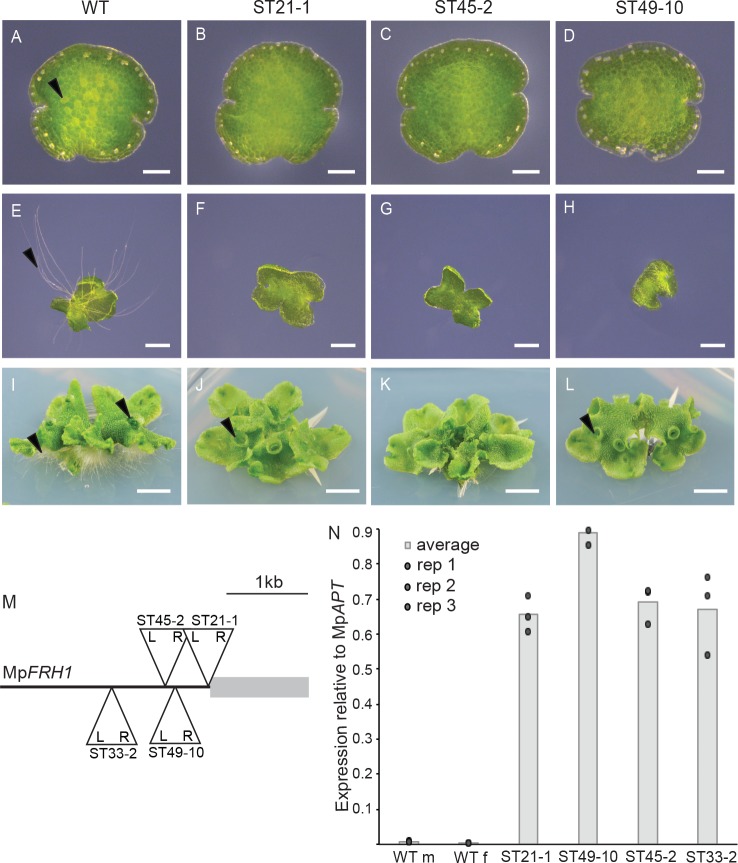
Figure 1. T-DNA insertion within the MpFRH1 promoter results in elevated steady state levels of MpFRH1 mRNA and defective rhizoid precursor cell differentiation.
(A–L) Phenotype of wild type M. polymorpha and the few rhizoid mutants ST21-1, ST45-2 and ST49-10. (A–D) One day old gemma (scale bar 100 μm), (E–H) four day old gemma (scale bar 1 mm), (I–L) 28 day old gemma (scale bar 5 mm) of wild type (A, E, I) and the few rhizoid mutants ST21-1 (N, F, J), ST49-10 (C, G, K) and ST45-2 (D, H, J). The arrowheads indicate rhizoid precursor cells (in A-D) rhizoids (in E-L) and gemma cups (in I-L). (M) Location and orientation of the T-DNA insertion sites within the MpFRH1 locus. L and R stand for T-DNA left and right border, respectively. (N) qRT-PCR analysis of steady state MpFRH1 mRNA levels in 15 day old gemmae of wild type and the few rhizoid mutants ST21-1, ST45-2 and ST49-10. The MpFRH1 transcript level was normalised against MpAPT1.