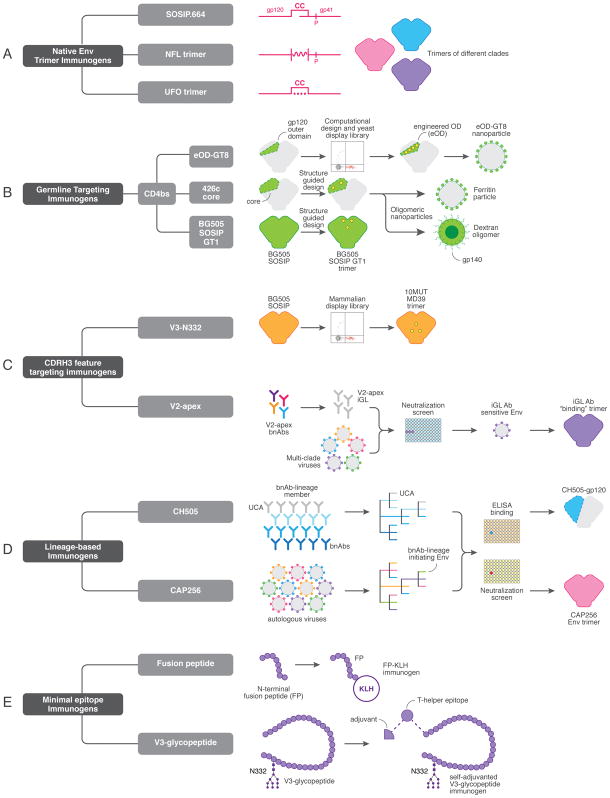
Figure 1. Immunogens that can form components of a neutralizing antibody-based HIV vaccine.
(a) Schematic showing native envelope trimer immunogens (SOSIP.664, native flexibly-linked (NFL) and uncleaved prefusion optimized (UFO) platforms), which form the basis of both priming and boosting immunization steps are shown. Trimers from multiple clades can be generated. (b) CD4 binding site (CD4bs) germline-targeting immunogens, including the engineered outer domain germline-targeting version 8 (eOD-GT8) immunogen (generated by computational approaches and refined through yeast-display and multimerized on nanoparticles), the 426c gp120 core, (which incorporates multiple glycan deletions around the CD4bs) and BG505 SOSIP-GT1 (SOSIP.664 modified to have enhanced binding of CD4bs germline-reverted antibodies) are illustrated. (c) Immunogens designed to select for antibodies with a long heavy chain complementarity determining region 3 (CDRH3), including the BG505 10MUT MD39 trimer to elicit PGT121-class V3-N332 bnAbs and trimers possessing “glycan holes” close to the trimer apex to generate V2 apex bnAbs. (d) Lineage-based immunogens, derived from virus-antibody co-evolution studies in donors CH505 and CAP256, are in development for the elicitation of CD4bs and V2 apex bnAbs. (e) Minimal epitope immunogens, including the N-terminal region of the fusion peptide (FP) fused to keyhole limpet hemocyanin (KLH) and V3-glycopeptides coupled to a T-helper epitope.