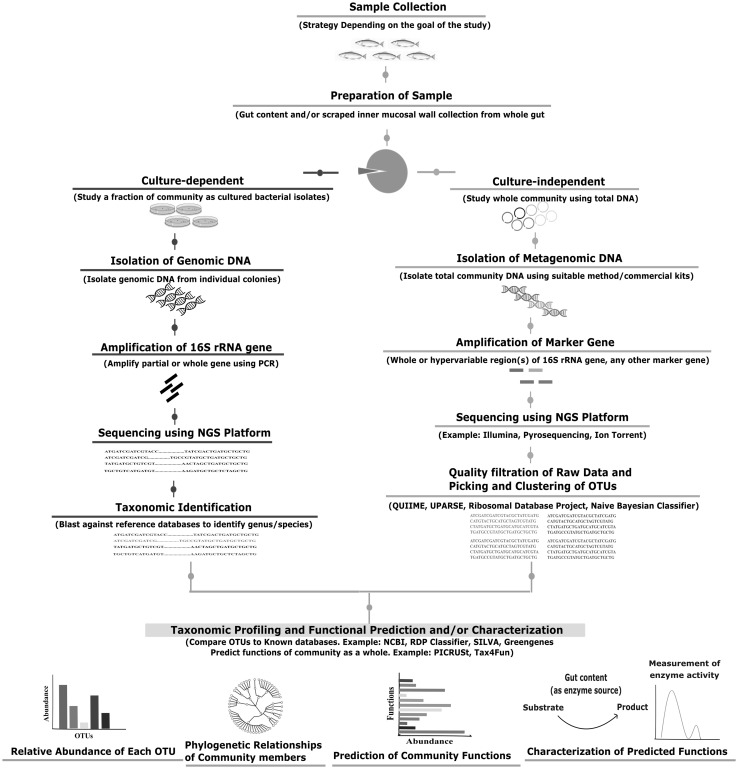
Fig. 2.
Schematic representation of the workflow for analyzing the fish gut microbiome. The currently employed procedures for fish gut microbiome studies include the traditional culture-dependent analysis as well as culture-independent analysis of the total DNA obtained directly from gut contents and mucosal wall. The culture-dependent techniques widely use sequencing of 16S rRNA gene to identify bacteria. For defining the uncultured microbiota, amplification and sequencing of whole or partial [hypervariable region(s)] of 16S rRNA gene is widely employed. Highly similar sequences are then grouped into Operational Taxonomic Units (OTUs) and compared against databases. The widely used databases and tools include NCBI [126], QIIME [127], UPARSE [128], Silva [129], Green Genes [130], RDP Classifier [131] and Naïve Bayesian Classifier [132]. The community is thus profiled based on the relative abundance of each OTU and their phylogenetic relationships. Using advanced ‘omic’ tools, functions of the community can also be predicted such as PICRUSt [34] or Tax4Fun [35]. Important microbial functions may then be characterized through wet lab experiments