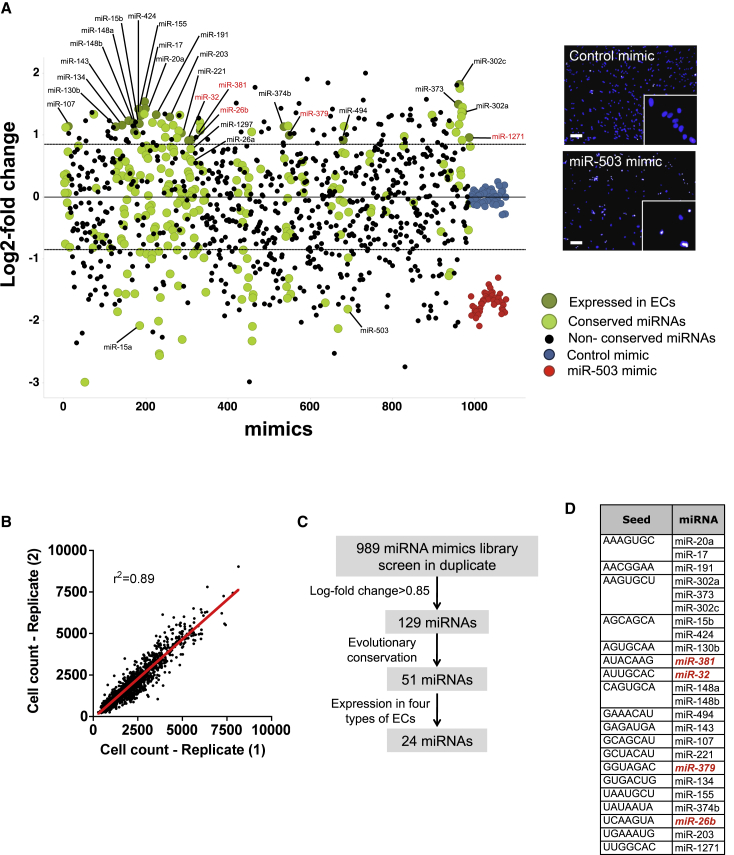
Figure 1.
Phenotypic Screen of miRNAs Regulating HUVEC Growth
(A) Left panel: log2 (fold change of miR mimics versus control mimics) values of cell count on the y axis is plotted against miR-mimics on the x axis. Non-conserved miRNAs (black), conserved miRNAs (light green), and miRNA enhancers of cell growth that are conserved and expressed in endothelial cells (dark green) are shown. Positive controls, miR-503 mimic, red; negative controls, miR-control mimic, blue; black dashed lines, cutoff log2 values in either direction >0.85 or <−0.85. Right panel: representative images of negative and positive controls from a screen plate. Nuclei were stained with DAPI (blue). Scale bar, 100 μm. (B) Correlation of screen plate replicates for the raw data of cell count parameter (r2 = 0.89). (C) Schematic describing the filtering and selection process of hit enhancers. (D) The selected hit enhancers are listed with their seed region. These miRNAs are expressed among four different EC types: HUVECs, human coronary artery endothelial cells (HCAECs), human aorta endothelial cells (HAECs), and human dermal micro-vascular endothelial cells (HMVECs). miRNAs that have not previously been studied in the context of angiogenesis are in red.