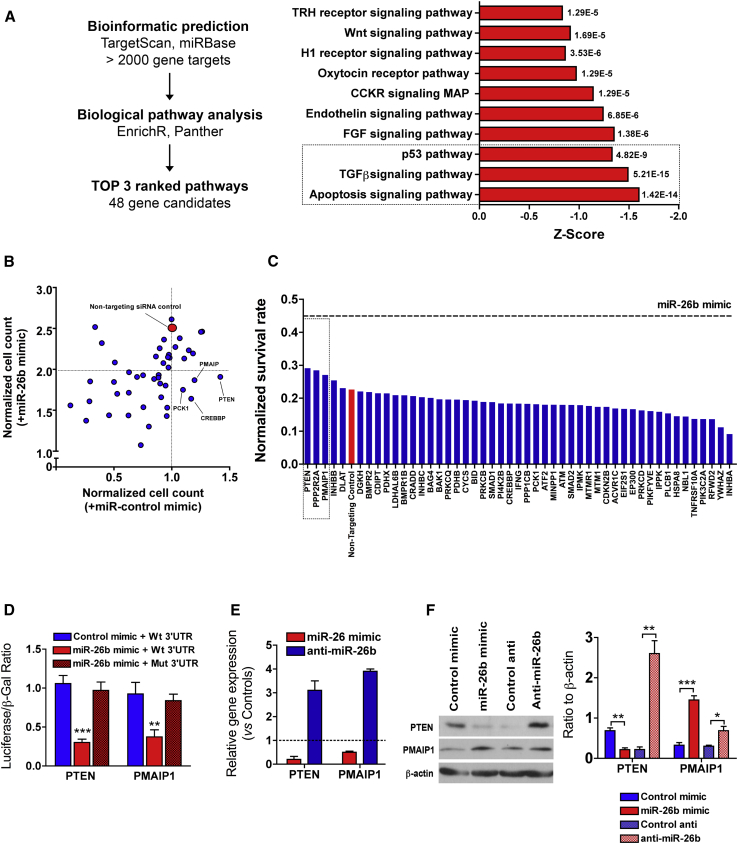
Figure 3.
Identification of miR-26b Target Genes
(A) Workflow for miRNA target identification (left panel). Genes were classified into molecular function according to the PANTHER classification system using EnrichR open source. Signaling pathways identified by the PANTHER classification system were plotted based on Z scores within each pathway (right panel). (B) siRNA-transfected HUVECs co-transfected with miR-control or miR-26b mimics. Cell count was captured 72 hr later. Cell count was normalized to non-targeting siRNA control-/miR-control mimic-transfected cells. Horizontal dashed line shows the cutoff value <2-fold for target genes abolishing the effect of pro-proliferative of miR-26b mimic. (C) siRNA-transfected HUVECs were incubated without or with a high dose of H2O2 (500 μM) for 24 hr. Horizontal dashed line shows the survival rate level of miR-26b mimic-transfected cells. Non-targeting siRNA control, red. (D) Luciferase gene reporter assays in HEK293 cells confirmed miR-26b binding to wild-type (WT) 3′ UTR or mutated (mut) 3′ UTR of PTEN and PMAIP1. Data are shown as the mean ± SEM of 6 independent experiments; **p < 0.01 and ***p < 0.001 versus miR-control mimic. Two-way ANOVA followed by Bonferroni post hoc test. (E) HUVECs were transfected with miR-26b mimic, anti-miR-26b, miR-control mimic, or anti-miR-control. At 3 days post-transfection, RNA was extracted and the levels of PTEN and PMAIP1 were determined by qRT-PCR. Values were normalized to S18 and then to the controls (mimic or anti-miR). Data are shown as the mean ± SEM of 4 independent experiments (*p < 0.01). (F) Cells were lysed and the expression of PTEN and PMAIP1 were analyzed by immunoblotting. β-actin was detected as a loading control.