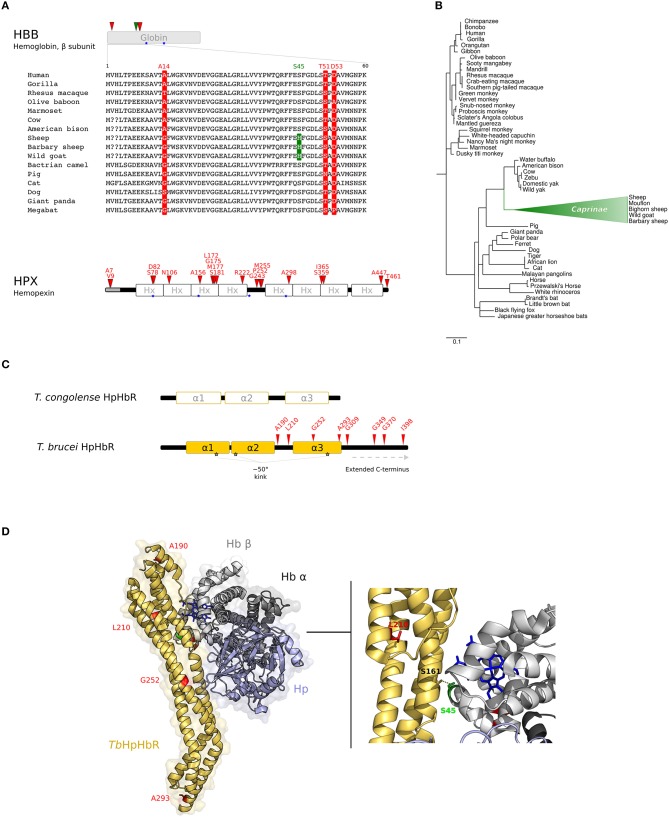
Figure 2.
Evolution at the interaction surface between Hb and Trypanosome brucei HpHbR. (A) Positively selected sites are mapped onto the β chain of Hb (HBB gene product) and on Hpx. A multiple alignment of the β chain of Hb (amino acids 1–60) for a few of representative mammalian species is shown. Sites that are positively selected in the mammalian phylogeny are marked in red; the site selected on the Caprinae branch is in green. Heme binding sites are indicated as blue stars. (B) aBS-REL analysis of positive selection for HBB in mammals. Branch lengths are scaled to the expected number of substitutions per nucleotide. The Caprinae branch is in green. (C) Schematic representation of trypanosome HpHbRs. Positively selected sites are shown in red. (D) 3D structure of human HpHb bound to TbHpHbR (PDB code: 5hu6); Hb is shown in gray (light, β subunit; dark, α subunit), Hp in light-blue, and HpHbR in light orange. Positively selected sites are mapped onto the structure; those that located at the contact interface are indicated in the enlargement. Hb-bound heme molecules are represented as blue sticks.