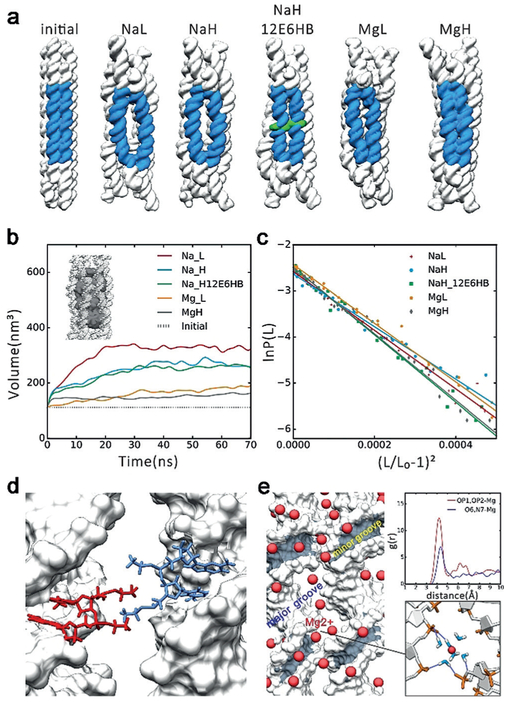
Figure 2.
MD simulation results. a) The initial conformations and averaged conformations of five systems in the last 10 ns. The big “O” rings are shown in blue, the ethyl-phosphorothioate-substituted DNA in green. NaL represents 6HB with 250 mM Na+, NaH represents 6HB with 500 mM Na+, NaH_12E6HB represents 6HB with 12 ethyl-phosphorothioated nucleotides with 500 mM Na+, MgL represents 6HB with 125 mM Mg2+, MgH represents 6HB with 250 mM Mg2+. b)Plot of inner volume vs. time for the center 42 bp of 6HBs. c)Stretch modulus of center 42 bp of 6HBs. d) Interaction model of 12E6HB, the hydrophobic ethyl group interacts in finger crossed conformation. e) Binding model of Mg2+ߝ6H2O. Left: Distribution of Mg2+ around helix (Mg2+ in red). Top right: Radial pair distribution function g(r) of Mg2+ around OP atoms and O6, N7 atoms of guanine. Bottom right: Bridging of the phosphates of two adjacent helixes by Mg2+–6H2O, the hydrogen bonds are shown in dark blue.