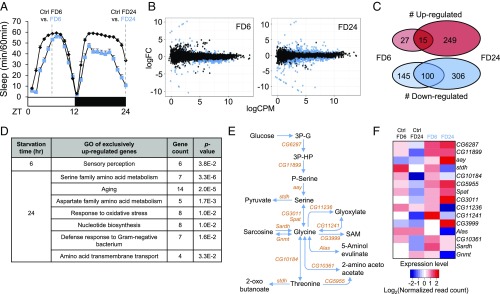
Fig. 1.
Transcriptome analyses of starved Drosophila brains identify up-regulation of serine biosynthesis pathway. (A) Schematic diagram depicting RNA-seq experimental design. Wild-type flies were fed 5% sucrose/1% agar (Ctrl) or deprived of sucrose for 6 and 24 h (FD6 and FD24). (B) Scatter plots demonstrate log counts per million (cpm) vs. log fold-change (FC) in expression of brains starved for 6 and 24 h. Blue dots represent DEGs (FDR < 0.05). (C) Venn diagram showing the number of genes that are regulated during short-term (FD6) and long-term (FD24) starvation. (D) Gene ontology analysis of genes that are up-regulated exclusively in FD6 and FD24. (E) Schematic diagram of the serine metabolic pathway. (F) Heat-map of expression level of genes involved in the serine metabolic pathway in fed (Ctrl FD6 and Ctrl FD24) and starved conditions (FD6 and FD24). Colors indicate the log2 values of normalized read counts.