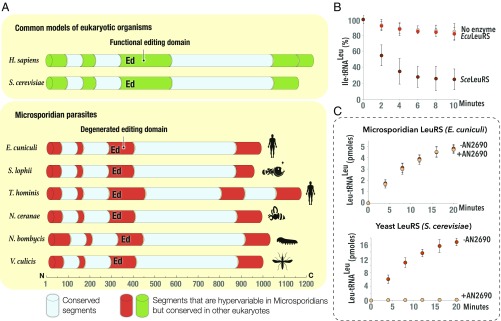
Fig. 2.
Microsporidian LeuRS synthetase lacks a proofreading domain and is naturally resistant to a drug from the benzoxaborole family. (A) Schematic structures of aminoacyl-tRNA synthetase LeuRS from different species, including several species of microsporidian parasites. The structures are color-coded by conservation: conserved segments are shown in blue (>50% sequence similarity between E. cuniculi and S. cerevisiae), segments degenerated in microsporidia but conserved in other eukaryotes are in green, and hypervariable segments in microsporidian LeuRS are in red (sequence similarity <35% between E. cuniculi and T. hominis). The scale indicates LeuRS length (in amino acids), and the silhouette symbols indicate a best-studied host (human, monkfish, honey bee, and silkworm) for each microsporidian parasite. The figure illustrates that, unlike LeuRS from other eukaryotic species, microsporidian LeuRS synthetases have a massively truncated editing domain, suggesting that microsporidian LeuRS synthetases lacks the editing activity. (B) Aminoacyl-tRNA hydrolysis assay to measure hydrolysis of the misaminoacylated substrate, Ile-tRNALeu, by microsporidian (E. cuniculi) or yeast (S. cerevisiae) LeuRS. The figure shows that unlike yeast LeuRS (used as a positive control), LeuRS from E. cuniculi is not capable of degrading the Ile-tRNALeu, illustrating a lack of the editing activity. (C) Kinetics of Leu-tRNALeu synthesis by microsporidian (E. cuniculi) and yeast (S. cerevisiae) LeuRS in the absence and in the presence of AN2690, a member of the benzoxaborole family that targets the editing domain of LeuRS synthetase. While both S. cerevisiae and E. cuniculi LeuRS synthetases are capable of producing Leu-tRNALeu in the absence of AN2690, only microsporidian LeuRS synthetase remains active in the presence of the drug, illustrating the natural resistance of E. cuniculi LeuRS to benzoxaboroles.