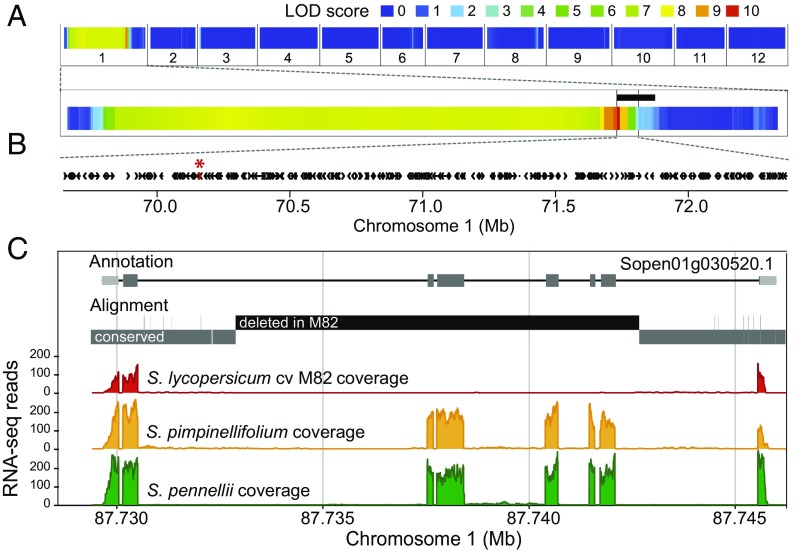
Fig. 1.
LNK2 colocalizes with the circadian period QTL and is partially deleted in cultivated tomato. (A) QTL analysis for circadian period in a S. pimpinellifolium x S. lycopersicum cv. MM RIL population identified a single significant locus on chromosome 1 (10). Logarithm of the odds (LOD) scores are given for the 12 tomato chromosomes; the genome-wide 5% significance threshold is 2.9. An introgression line with a precisely defined genomic fragment from the wild tomato relative S. pennellii (indicated by a black bar) exhibited the same short-period phenotype (SI Appendix, Fig. S1) and refined the left border of the QTL. (B) The 245 genes annotated in the tomato genome reference v2.4 present in the QTL region are depicted by black arrows; genes deleted in cultivated tomato but present in S. pennellii are shown in red and marked by an asterisk. (C) From top to bottom: gene model for LNK2 in the wild tomato species S. pennellii; graphical output of BLAST results comparing the genomic sequence of S. pennellii and cultivated tomato; and coverage plots of RNA-seq reads from cultivated tomato (red), S. pimpinellifolium (orange), and S. pennellii (green), aligned to the S. pennellii reference genome.