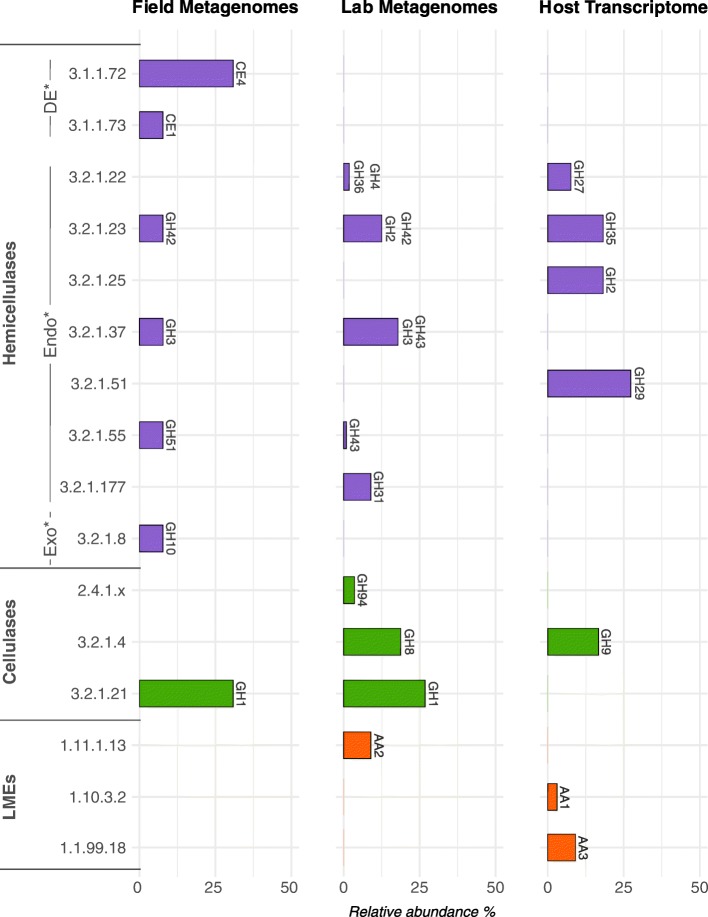
Fig. 2.
Prediction of enzymatic functions (EC number) of debranching enzymes (DE), endo-hemicellulases (Endo), exo-hemicellulases (Exo), cellulases, and lignin modifying enzymes (LMEs) identified in the metagenomes of specimens from the field and the laboratory and in the host transcriptome. Relative abundance (in %) for a given predicted enzymatic function was calculated by dividing the identified counts for a given enzyme by the total counts identified in a metagenome or in the transcriptome