Figure 1. Summary of potential clinical applications of ctDNA sequencing throughout different stages of cancer management.
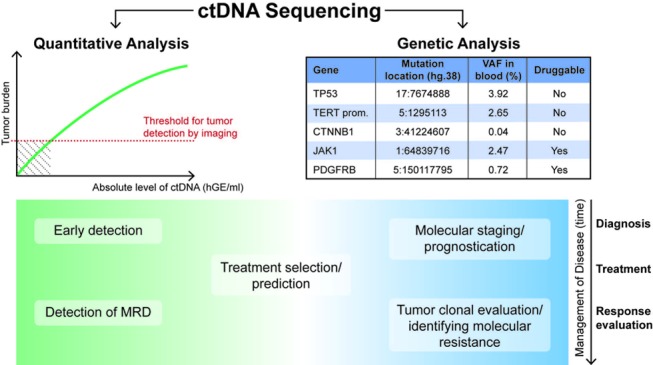
Quantitative analysis of ctDNA is based on a positive correlation of ctDNA concentration (e.g. haploid genome equivalents per mL, hGE/mL) and (absolute or metabolic) tumor burden [4,7], and thus potentially able to overcome a limiting detection threshold of imaging for very small tumors (hatched area). For example, in the setting of detection of minimal residual disease (MRD) after surgical and/or locoregional therapies. In contrast, analysis of specific genetic (e.g. mutations, see table) [3] or epigenetic (e.g. DNA methylation) aberrations could allow to identify targets for therapies and monitor how clonal composition evolves over time and upon exposures to treatments [6,7]. This might facilitate real time identification of molecular mechanisms of tumor resistance.
