Abstract
Trichogramma is a kind of egg parasitoid wasp that is widely used to control lepidopterous pests. Temperature is one of the main factors that determines the various life activities of this species, including development, reproduction and parasitism efficiency. Heat shock proteins (HSPs) are highly conserved and ubiquitous proteins that are best known for their responsiveness to temperature and other stresses. To explore the potential role of HSPs in Trichogramma species, we obtained the full-length cDNAs of six HSP genes (Tchsp10, Tchsp21.6, Tchsp60, Tchsp70, Tchsc70-3, and Tchsp90) from T. chilonis and analyzed their expression patterns during development and exposure to temperature stress. The deduced amino acid sequences of these HSP genes contained the typical signatures of their corresponding protein family and showed high homology to their counterparts in other species. The expression levels of Tchsp10, Tchsp21.6 and Tchsp60 decreased during development. However, the expression of Tchsc70-3 increased from the pupal stage to the adult stage. Tchsp70 and Tchsp90 exhibited the highest expression levels in the adult stage. The expression of six Tchsps was dramatically upregulated after 1 h of exposure to 32 and 40°C but did not significantly change after 1 h of exposure to 10 and 17°C. This result indicated that heat stress, rather than cold stress, induced the expression of HSP genes. Furthermore, the expression of these genes was time dependent, and the expression of each gene reached its peak after 1 h of heat exposure (40°C). Tchsp10 and Tchsp70 exhibited a low-intensity cold response after 4 and 8 h of exposure to 10°C, respectively, but the other genes did not respond to cold at any time points. These results suggested that HSPs may play different roles in the development of this organism and in its response to temperature stress.
Introduction
Wasps of the genus Trichogramma (Hymenoptera: Trichogrammatidae) are tiny egg parasitoids of numerous insect species that are distributed around the world [1–3]. These wasps are easily mass reared and have a broad range of hosts [4]. Since 1975, several species of Trichogramma have been commonly used as biological control agents for various pests in agricultural and forest systems [5, 6]. Among these parasitoid wasps, T. chilonis is one of the most successful species in controlling lepidopterous pests, including Chilo spp. (Lepidoptera: Pyralidae), Helicoverpa armigera (Lepidoptera: Noctuidae) and Pectinophora gossypiella (Lepidoptera: Gelechiidae) [2]. In China, T. chilonis is widely distributed and is employed in integrated pest management for rice, cotton, sugarcane and other crops [7].
Temperature is a vital factor that determines the distribution and abundance of animals [8]. It is also crucial for the successful introduction of Trichogramma species because it influences their development, survival, reproduction, and sex ratio, as well as their parasitism efficiency [3, 9]. Previous studies have claimed that Trichogramma species can live under a wide range of temperatures from 9 to 36°C [10]. The temperature range of 25–30°C is considered optimal for rearing T. chilonis in the laboratory [11]. Temperatures beyond the optimal conditions could cause detrimental effects on various biological aspects of the wasps. For instance, T. chilonis and three other Trichogramma species cannot parasitize the eggs of Cnaphalocrocis medinalis (Guene´e) (Lepidoptera: Pyralidae) at 36°C [12]. The emergence and host parasitization of T. chilonis are 98.0% and 95.6% at 28°C but decrease to 33.7% and 60.1%, respectively, at 35°C [9]. Moreover, a low temperature of 15°C leads to long developmental periods for T. chilonis (26.3 days) and Trichogrammatoidea bactrae (25.6 days) (Hymenoptera: Trichogrammatidae) [4]. Although detrimental consequences caused by temperature stress have been well reported, little is known about the molecular response to temperature stress in Trichogramma.
Heat shock proteins (HSPs) are highly conserved and ubiquitous proteins that are best known for their responsiveness to multiple stresses such as extreme temperatures, desiccation, anoxia, hypertonic stress, ultraviolet radiation, heavy metals, ethanol, and other contaminants [13, 14]. In general, HSPs act as molecular chaperones that promote correct refolding of proteins and prevent the misfolding or aggregation of proteins [15]. According to their molecular weight and homology, HSPs are classified into several families, including HSP100, HSP90, HSP70, HSP60, HSP40 and small HSPs (sHSPs) [16]. To data, certain groups of HSP genes (hsps) have been identified and cloned from various insects such as Spodoptera litura (Lepidoptera: Noctuidae), Thitarodes pui (Lepidoptera: Hepialidae), Leptinotarsa decemlineata (Coleoptera: Chrysomelidae) and C. suppressalis [17–20]. An increasing number of hsps have been shown to respond to temperature stress [17, 21, 22]. In an endoparasitoid wasp, Pteromalus puparum (Hymenoptera: Pteromalidae), six hsps are induced by 1 h of exposure to −3 and 36°C, including hsp20, hsp40, hsp60, hsp70, hsc70, and hsp90 [23]. The expression patterns of five hsps vary but are indeed induced by heat or cold stress in Cotesia chilonis (Hymenoptera: Braconidae) [24]. In addition, hsps have also been reported to be involved in the developmental processes of endoparasitoid wasps such as Venturia canescens (Hymenoptera: Ichneumonidae), Macrocentrus cingulum (Hymenoptera: Braconidae) and C. vestalis [25–27].
Trichogramma wasps are often released in the fields during pupal stage [5]. Various biological parameters of the released parasitoids are influenced by ambient temperature [3, 9]. To data, the molecular mechanism of thermal tolerance remains unclear. In present study, six hsps of T. chilonis (Tchsp10, Tchsp21.6, Tchsp60, Tchsp70, Tchsc70-3, and Tchsp90) were cloned and characterized, and their expression profiles during development were explored. In addition, individuals at the pupal stage were collected to explore the expression patterns of these six hsps in response to various levels of temperature stress (10, 17, 32 and 40°C for 1 h). The temporal expression patterns of six Tchsps were also investigated during cold (10°C) and heat (40°C) exposure. To our knowledge, this is the first report on the isolation and analysis of hsps from T. chilonis. Our results are expected to help elucidate the potential contribution of these HSPs to thermal tolerance and development.
Materials and methods
Insects
Prepupae of T. chilonis (parasitized eggs) and eggs of Corcyra cephalonica (Stainton) (Lepidoptera: Pyralidae) were obtained from the Plant Protection Research Institute, Guangdong Academy of Agricultural Sciences, People’s Republic of China. T. chilonis cultures were maintained on irradiated C. cephalonica eggs for several generations at 25 ± 1°C with 75 ± 5% relative humidity and a 14 L:10 D photoperiod.
Sampling at different developmental stages
The irradiated eggs of C. cephalonica were glued on 4 paper cards (2 × 1 cm) and exposed to freshly emerged T. chilonis for 30 min. These egg cards were transferred to different glass cylinders and maintained at 25 ± 1°C with 75 ± 5% relative humidity and a 14 L:10 D photoperiod. Parasitized eggs on different cards were dissected to collect the larvae, prepupae, pupae and adults of T. chilonis. The developmental stages of T. chilonis were confirmed under a stereoscope as described in previous studies [28, 29]. Every 6 h, a small number of parasitized eggs were dissected to determine the developmental stage of T. chilonis. At the larval stage of T. chilonis, the colors of individuals and parasitized eggs were both white. Larvae with oval shapes were collected. At the prepupal stage, the color of parasitized eggs was black, and pulm spots were visible on the body. Prepupae were collected when pulm spots disappeared from the head and tail. At the pupal stage, the color of parasitized eggs turned deep, red compound eyes appeared on the body, and pulm spots disappeared. Pupae with small black spots on their bodies were collected. The adults were collected once they emerged from the eggs. To collect corresponding individuals, parasitized eggs were immediately placed on a filter-paper soaked with Sample Protector for RNA/DNA (TaKaRa, Dalian, China) and dissected under a stereoscope. The specimens were immediately transferred to TRIzol (Invitrogen, Darmstadt, Germany) and stored in a -80°C refrigerator. Fifty wasps from each developmental stage were collected. The experiment was repeated three times.
Temperature exposure
Considering that T. chilonis wasps are often released as pupae inside the host eggs, these parasitized eggs were chosen for temperature exposure experiments. The parasitized eggs were exposed to temperatures of 10, 17, 32 and 40°C for 1 h, and parasitized eggs kept at 25°C were collected as controls. These parasitized eggs were then dissected to collect wasps under a stereoscope. In addition, wasps were collected at different time points (1, 2, 4 and 8 h) during cold (10°C) and heat (40°C) exposure. The sampling method was the same as described above.
Cloning the full-length cDNA of hsps
Total RNA from adults was isolated with a TRIzol Reagent Kit according to the supplier’s instructions. Assessment of the quality and quantity of total RNA was performed by electrophoresis and with a NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, DE, USA). First-strand cDNA was generated with a PrimeScriptTM RT Reagent Kit (TaKaRa, Dalian, China). Templates for 5’ and 3’ RACE were constructed using a SMARTTM RACE cDNA Amplification Kit (Clontech, California, USA). Primers (S1 Table) were designed based on the nucleotide sequences from the transcriptome data of T. chilonis (SRA accession number: SRP119024). PCR products were cloned and then sequenced by Sangon (Shanghai).
Bioinformatics analysis
Using the DNASTAR software package, full-length cDNAs of hsps were obtained based on the sequenced fragments. The BLAST search was performed to find homologous sequences in GenBank. Multiple sequence alignment and identity analysis were performed using DNAMAN software. The open reading frame (ORF) and deduced amino acid sequence of each hsp were identified and obtained using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The predicted molecular weight and theoretical isoelectric point (pI) of the deduced proteins were predicted with the ExPASy (http://www.expasy.org/). Domains were predicted by SMART tool (http://smart.embl-heidelberg.de/). Phylogenetic analysis was performed using MEGA software (version 6.0) with the 1000 bootstrap replicates. Five neighbor-joining (NJ) phylogenetic trees were constructed using members of HSP10, sHSPs, HSP60, HSP70 and HSP90 familiy.
Quantitative real-time PCR
Total RNA of the samples from each treatment was extracted and reverse transcribed as described above. Primers were designed based on the conserved regions of the hsps, and glyceraldehyde-3-phosphate dehydrogenase (gapdh) was used as the control (S1 Table). The real-time PCR reaction was performed in a 10 μL reaction volume following the manufacturer’s protocol for SYBR ® Premix Ex Taq™ (TaKaRa, Dalian, China). The expression profiles of hsps were determined on a Roche 480 Real-Time PCR System (Roche, Switzerland) under the following conditions: 95°C for 3 min, 40 cycles of 95°C for 10 s, 60°C for 20 s and 72°C for 20 s. The melting curve analysis was applied to ensure the specificity of primers at the end of the program. The relative abundance of each hsp was calculated according to the 2−ΔΔCt method [30].
Statistics
The expression values of the hsps are presented as the means ± SEM. Statistical analysis was performed by SPSS v.16.0 software (SPSS, Chicago, IL, USA) with one-way analysis of variance (ANOVA) and Duncan’s post hoc tests.
Results
Characterization of hsp genes
Tchsp10.
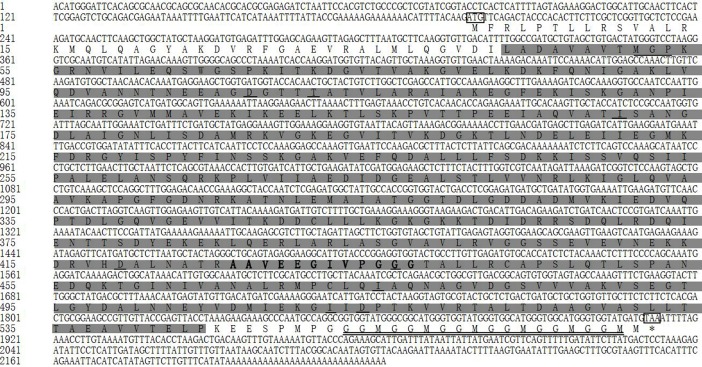
The full-length cDNA of Tchsp10 was 705 bp, including an ORF of 315 bp, a 5’- untranslated region (UTR) of 205 bp and a 3’-UTR of 185 bp (GenBank accession number MH490973). The ORF of Tchsp10 encoded a polypeptide of 104 amino acids with a predicted molecular weight of 11.26 kDa and a pI of 8.93. TcHSP10 showed topical Cpn10 superfamily characteristics with a conserved domain (aa 9–102) and and a mobile loop (aa 25–38) (Fig 1).
Fig 1. Nucleotide sequence and deduced amino acid sequence of Tchsp10.
The initiation and stop codons are marked with boxes. The conserved domain is shaded in light gray. The mobile loop is underlined.
Tchsp21.6.
The full-length cDNA of Tchsp21.6 was 2119 bp, including an ORF of 576 bp, a 5’- UTR of 218 bp and a 3’-UTR of 1325 bp (GenBank accession number MH490974). The ORF of Tchsp21.6 encoded a polypeptide of 191 amino acids with a predicted molecular weight of 21.68 kDa and a pI of 5.6. TcHSP21.6 was a typical small HSP, containing a metazoan α-crystalline domain (ACD) (Fig 2).
Fig 2. Nucleotide sequence and deduced amino acid sequence of Tchsp21.6.
The initiation and stop codons are marked with boxes. The α-crystallin domain (ACD) is shaded in light gray.
Tchsp60
The full-length cDNA of Tchsp60 was 2222 bp, including an ORF of 1716 bp, a 5’-UTR of 197 bp and a 3’-UTR of 309 bp (GenBank accession number MH490975). The ORF of Tchsp60 encoded a polypeptide of 571 amino acids with a predicted molecular weight of 60.48 kDa and a pI of 5.18. TcHSP60 contained a classical mitochondrial HSP60 signature motif (AAVEEGIVPGGG), a C-terminal Gly-Gly-Met repeat (GGM repeat motif) and ATP/ADP binding sites (Fig 3).
Fig 3. Nucleotide sequence and deduced amino acid sequence of Tchsp60.
The initiation and stop codons are marked with boxes. The conserved domain is shaded in light gray. The ATP binding sites are underlined. The GGM repeat motif is marked with a double line. The classical mitochondrial HSP60 signature motif is shown in bold.
Two TcHSP70 genes
The full-length cDNA of Tchsp70 was 2573 bp, including an ORF of 1992 bp, a 5’-UTR of 13 bp and a 3’-UTR of 568 bp (GenBank accession number MH490976). The ORF of Tchsp70 encoded a polypeptide of 663 amino acids with a predicted molecular weight of 73.26 kDa and a pI of 5.62.
The full-length cDNA of Tchsc70-3 was 2668 bp, including an ORF of 1992 bp, a 5’-UTR of 186 bp and a 3’-UTR of 490 bp (GenBank accession number MH490977). The ORF of Tchsc70-3 encoded a polypeptide of 663 amino acids with a predicted molecular weight of 73.34 kDa and a pI of 5.12.
The two TcHSP70 sequences contained three conserved signatures, an ATP-GTP binding site and a non-organellar consensus motif (Fig 4). In addition, the KDEL motif was identified in the deduced amino acid sequence of Tchsc70-3. The EEVD motif was found at the C-terminus of TcHSP70.
Fig 4. Multiple amino acid sequence alignments of two genes in the HSP70 family.
Three signature motifs of the HSP70 family are shown in light gray. The non-organellar consensus motif is boxed, the localization motif is underlined, and the ATP/GTP binding site is double underlined.
Tchsp90
The full-length cDNA of Tchsp90 was 2643 bp, including an ORF of 2181 bp, a 5’-UTR of 145 bp and a 3’-UTR of 317 bp (GenBank accession number MH490980). The ORF of Tchsp90 encoded a polypeptide of 726 amino acids with a predicted molecular weight of 83.48 kDa and a pI of 4.88.
Five highly conserved signature sequences of the HSP90 family were found, including NKEIFLRELISNSSDALDKIR (aa 41–61), LGTIAKSGT (aa 108–116), IGQFGVGFYSAYLVAD (aa 132–147), IKLYVRRVFI (aa 357–366) and GVVDSEDLPLNISRE (aa 383–397) (Fig 5). The MEEVD motif was identified at the C-terminus of the deduced amino acid sequence.
Fig 5. Nucleotide sequence and deduced amino acid sequence of Tchsp90.
The initiation and stop codons are marked with boxes. Five signature motifs of the HSP90 family are shown in light gray. The localization motif is double underlined.
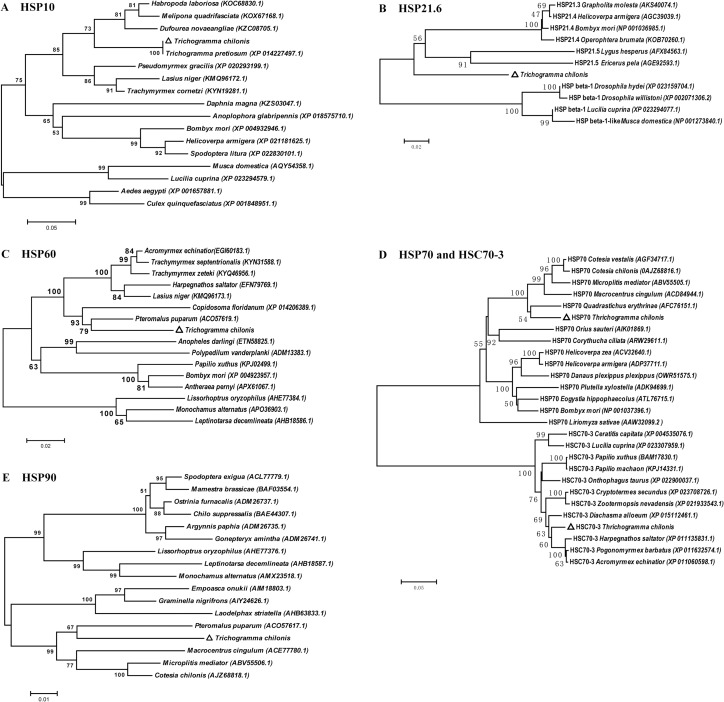
Phylogenetic analysis of TcHsps
Phylogenetic trees were constructed based on the deduced amino acid sequences of Tchsps and their homologous sequences by the neighbor-joining method. The results revealed that HSP10 sequences from T. chilonis and T. pretiosum were clustered together into a single branch (Fig 6(A)). Two HSP60 sequences of endoparasitoid wasps (T. chilonis and P. puparum) were clustered within a branch (Fig 6(C)). A similar result was also found in the phylogenetic tree constructed with HSP90 sequences (Fig 6(E)). TcHSP21.6 showed a high similarity with HSP21.5 and HSP21.4 from other insects (Fig 6(B)). These sHSPs were clustered together and were separated from HSP beta-1 sequences. The sequences of the HSP70 family from insects presented two clusters, one with HSP70 sequences and another with HSC70-3 sequences (Fig 6(D)). The two TcHSP70 sequences showed a close relationship with their homologous sequences from the hymenopteran species.
Fig 6. Phylogenetic analysis of TcHSPs and other homologous sequences from insects.
The Neighbor-Joining (NJ) trees are constructed by using MEGA 6.0. The positions of HSPs of Trichogramma chilonis are marked with triangles. (A) HSP10, (B) small HSP, (C) HSP60, (D) HSP70 and HSC70-3, and (E) HSP90.
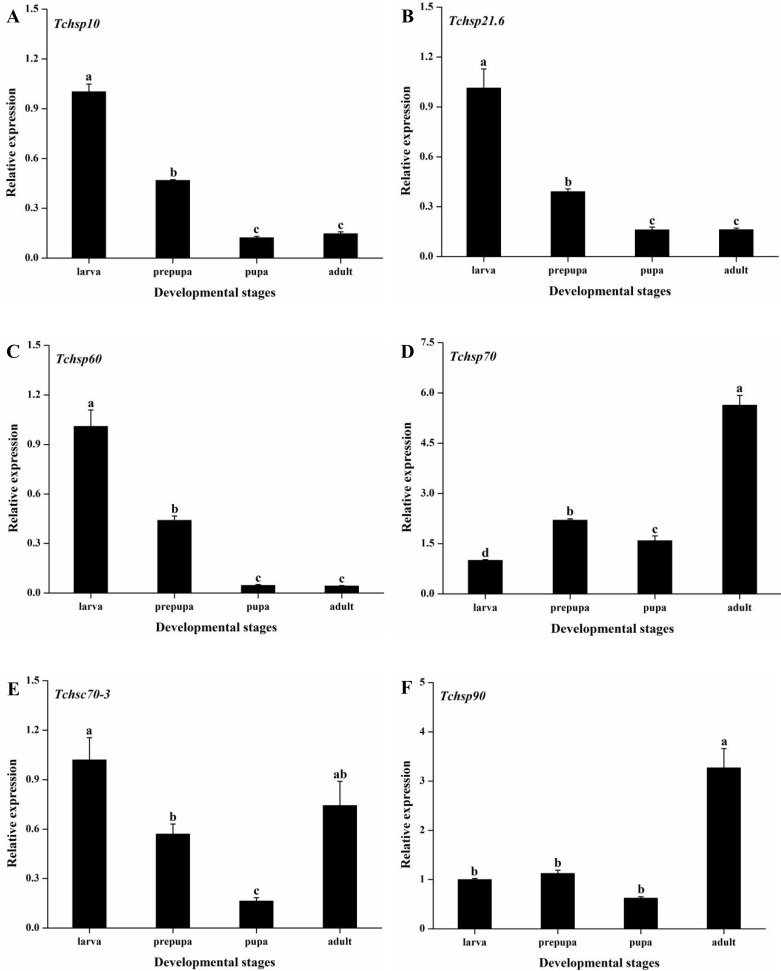
Expression of Tchsps during development
Real-time PCR was used to measure the expression levels of Tchsps during development. The expression level in the larval stage was used as the control value. The developmental expression profiles of six Tchsps varied significantly in T. chilonis (Tchsp10: F3, 8 = 304.50, p < 0.001; Tchsp21.6: F3, 8 = 98.80, p < 0.001; Tchsp60: F3, 8 = 332.64, p < 0.001; Tchsp70: F3, 8 = 183.91, p < 0.001; Tchsc70-3: F3, 8 = 11.67, p = 0.003; Tchsp90: F3, 8 = 115.27, p < 0.001). The expression of Tchsp10, Tchsp21.6 and Tchsp60 decreased from the larval stage to the pupal stage and was maintained at a low level during the pupal and adult stages (Fig 7). Similarly, Tchsc70-3 had the highest expression level during the larval stage and the lowest level during the pupal stage, while it was upregulated from the pupal stage to the adult stage. In contrast, the expression of Tchsp70 and Tchsp90 peaked in the adult stage, with 5.63- and 3.27-fold increases, respectively compared to the levels in the larval stage.
Fig 7. Relative expression levels of Tchsps during development.
Data are presented as the means ± SE (n = 3). Different lowercase letters indicate significant differences. (A) Tchsp10, (B) Tchsp21.6, (C) Tchsp60, (D) Tchsp70, (E) Tchsc70-3 and (F) Tchsp90.
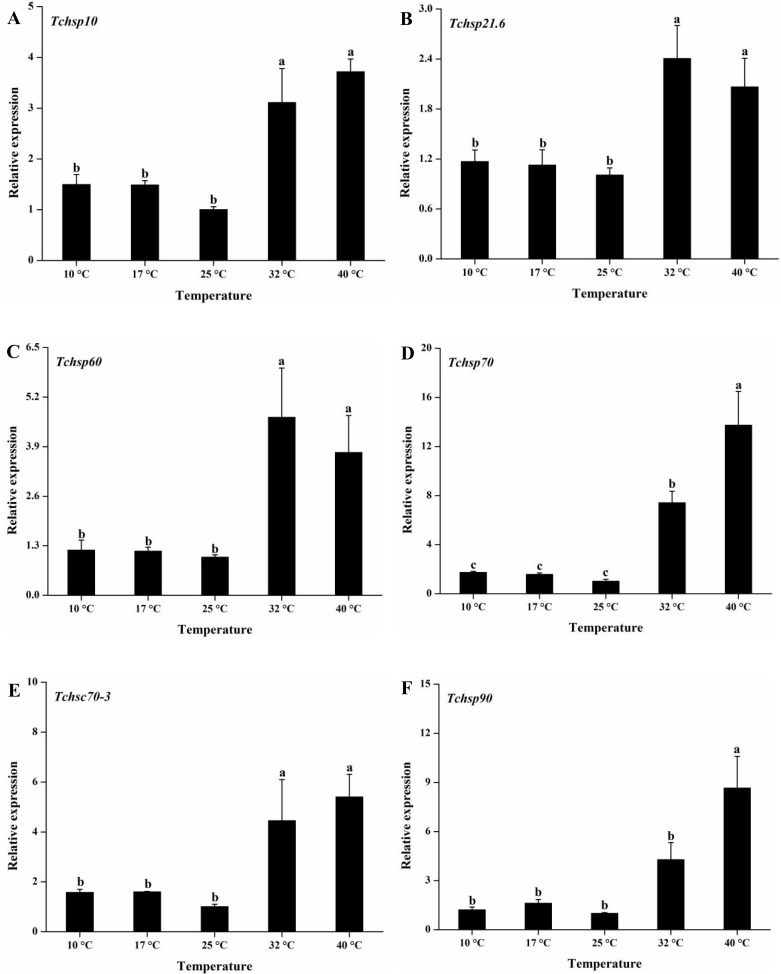
Expression profiles of Tchsps at different temperatures
All six Tchsps showed the same expression pattern after exposure to different temperatures (10, 17, 25, 32 and 40°C) for 1 h (Fig 8). They were all significantly upregulated at high temperatures (32 and 40°C) compared with 25°C (Tchsp10: F4, 10 = 20.71, p < 0.001; Tchsp21.6: F4, 10 = 6.04, p = 0.01; Tchsp60: F4, 10 = 5.56, p = 0.013; Tchsp70: F4, 10 = 64.07, p < 0.001; Tchsc70-3: F4, 10 = 24.07, p < 0.001; Tchsp90: F4, 10 = 22.91, p < 0.001). Although the expression of Tchsp90 at 32°C was higher than the levels at 10, 17, and 25°C, there were no significant differences. The expression levels of Tchsp70, Tchsc70-3 and Tchsp90 were significantly increased from 32 to 40°C. Among these genes, Tchsp70 had the greatest heat response, with a 7.41 -fold increase at 32°C and a 13.74-fold increase at 40°C. On the other hand, the expression levels of six Tchsps slightly increased after exposure to 10 and 17°C but were not significantly different from the expression levels at 25°C.
Fig 8. Relative expression levels of Tchsps after 1 h of exposure to different temperatures.
Data are presented as the means ± SE (n = 3). Different lowercase letters indicate significant differences. (A) Tchsp10, (B) Tchsp21.6, (C) Tchsp60, (D) Tchsp70, (E) Tchsc70-3 and (F) Tchsp90.
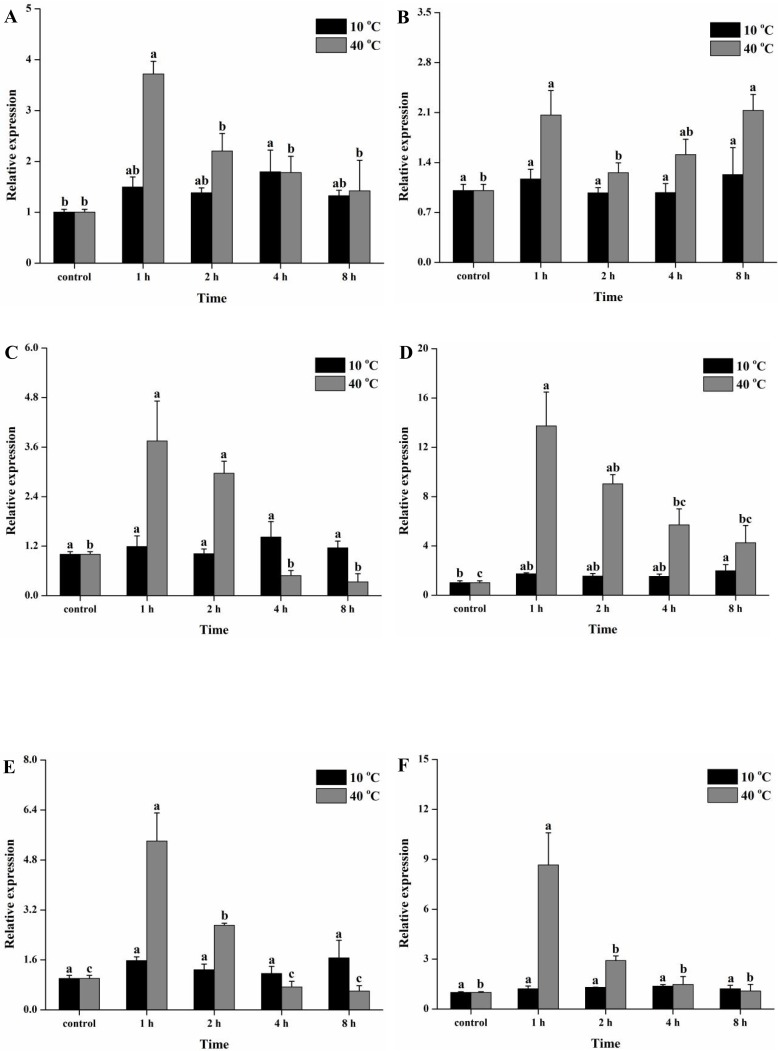
The temporal expression patterns of six Tchsps were also investigated during cold (10°C) and heat (40°C) exposure. The results indicated that the expression levels of Tchsp10 and Tchsp70 were significantly increased after 4 and 8 h of cold exposure (10°C), respectively (F4, 10 = 3.48, p ≤ 0.05; F4, 10 = 3.69, p = 0.043). Although the expression levels of other Tchsps were also slightly increased after cold exposure for different periods of time, they showed no significant differences compared with the control group (Tchsp21.6: F4, 10 = 0.31, p = 0.87; Tchsp60: F4, 10 = 0.49, p = 0.74; Tchsc70-3: F4, 10 = 1.37, p = 0.31; Tchsp90: F4, 10 = 1.20, p = 0.37) (Fig 9). On the other hand, the six Tchsps were strongly expressed after 1 h of heat exposure (40°C) (Tchsp10: F4, 10 = 10.37, p = 0.001; Tchsp21.6: F4, 10 = 5.08, p = 0.017; Tchsp60: F4, 10 = 11.19, p = 0.001; Tchsp70: F4, 10 = 9.98, p = 0.002; Tchsc70-3: F4, 10 = 23.38, p < 0.001; Tchsp90: F4, 10 = 12.25, p = 0.001). The expression of Tchsp10, Tchsp60, Tchsp70, Tchsp70-3 and Tchsp90 decreased from 1 h to 8 h. However, Tchsp21.6 exhibited the highest levels at 1 h and 8 h.
Fig 9. Temporal expression patterns of Tchsps during cold (10°C) and heat (40°C) exposure.
Data are presented as the means ± SE (n = 3). Different lowercase letters indicate significant differences. (A) Tchsp10, (B) Tchsp21.6, (C) Tchsp60, (D) Tchsp70, (E) Tchsc70-3 and (F) Tchsp90.
Discussion
In this study, full-length cDNAs of six HSP genes were obtained from T. chilonis, including Tchsp10, Tchsp21.6, Tchsp60, Tchsp70, Tchsc70-3 and Tchsp90. TcHSP10 contained a mobile loop, which is consistent with the characteristic of other HSP10 sequences described in many studies [31–33]. Through the mobile loop, HSP10 interacts with the HSP60, which helps the folding of protein [34, 35]. Tchsp21.6 encoded a polypeptide of 191 amino acids with a predicted molecular weight of 21.68 kDa. The deduced amino acid sequence of Tchsp21.6 contained an α-crystallin domain (ACD), which is a characteristic feature of the sHSPs family [36, 37]. This family is composed of many members containing variable N- and C-terminal extensions [32, 38]. TcHSP60 belongs to the mitochondrial HSP60 family, containing a conserved signature motif (AAVEEGIVPGGG) and a GGM motif [23, 39]. The GGM motif at the C-terminus has been suggested to provide a suitable physical environment for protein folding [40]. The ATP/ADP binding sites were found in TcHSP60, which have also been identified in HSP60 sequences from Rhopalosiphum padi (L.) (Homoptera: Aphididae) and Lucilia cuprina (Diptera: Calliphoridae) [39, 41]. The highly conserved motif among HSP60 sequences may indicate that a similar mechanism of coupling ATP hydrolysis to the substrate-refolding process exists [14, 39, 42]. The two TcHSP70 sequences had three conserved HSP70 family signatures and a non-organellar consensus motif, in accordance with the structures of the HSP70 sequences described in Nilaparvata lugens (Homoptera: Delphacidae), Sitodiplosis mosellana (Diptera: Cecidomyiidae) and Habrobracon hebetor (Hymenoptera: Braconidae) [43–45]. The two TcHSP70 sequences showed high similarity with their homologous sequences from other insects. These results indicated that the members of HSP70 family are highly conserved [46].
It has been well reported that HSPs are involved in the development of insects [47, 48]. However, the expression patterns of hsps during development vary in insects [49, 50]. For example, the expression of hsp60 increases from the larval stage to the adult stage in Liriomyza sativa (Diptera: Agromyzidae), while it decreases from nymph to adult in R. padi [39, 47]. In this study, Tchsp10 and Tchsp60 levels decreased during development, which is consistent with the findings of report on Galeruca daurica (Coleoptera: Chrysomelidae) [50]. Moreover, Tchsp21.6 showed the same expression pattern as Tchsp10 and Tchsp60. The high expression of three Tchsps in the larval stage indicated that these genes may be related to larval development. In T. chilonis, Tchsp70 and Tchsp90 levels peaked at the adult stage, and Tchsc70-3 expression was upregulated from the pupal stage to the adult stage. It is different from the findings in S. exigua and Frankliniella occidentalis (Thysanoptera: Thripidae) [48, 49]. In contrast, in the endoparasitoid wasp M. cingulum, hsp70 and hsp90 are highly expressed in the pupal and adult stages [26]. The high expression of hsp70 begins at the third-instar larval stage, when C. vestalis comes out of the host [27]. As an egg endoparasitoid, adults of T. chilonis emerge from host, thus facing very different environmental stresses. These HSP genes, i.e., Tchsp70, Tchsp90 and Tchsc70-3, might be needed to overcome these challenges.
HSPs also play important roles in the response to temperature stress [51]. As described in the introduction, HSPs are molecular chaperones that help to prevent potential damage to cellular and molecular structures under temperature and other stresses [15, 52]. Altered expression patterns of hsps have been widely reported under temperature stresses, although these responses seem to be species-specific among insects [39, 43]. In T. pui, the expression of hsp90, rather than hsp70, changes in response to temperature [22]. However, hsp90 and hsp70 in Empoasca onukii (Hemiptera: Cicadellidae) are both highly expressed under cold and heat treatments [53]. Cchsp60 in C. chilonis responds to cold stress but is insensitive to heat stress [24]. In contrast, the highest expression level of hsp60 in P. puparum appears at 36°C [23]. In this study, Tchsps were sensitive to high temperatures (32 and 40°C), which is consistent with the expression patterns of hsps observed in other species [23, 48, 50]. The expression of Tchsp10, Tchsp21.6 and Tchsp60 showed the same expression pattern in response to high temperatures, each being significantly upregulated under high temperatures but with no significant differences at 32 and 40°C. The upregulation of hsp10 and hsp60 has also been reported in Apostichopus japonicus (Echinodermata: Holothuroidea) and G. daurica [31, 50]. HSP10 is considered the co-chaperone of HSP60 and interacts with HSP60 in the same heat shock pathway [33, 54]. TcHSP21.6 belongs to the sHSPs family and showed high similarity with HSP21.5 and HSP21.4, which have been proven to respond to heat stress in E. pela female adults and in B. mori [55, 56]. Tchsp70 exhibited the highest expression at 40°C. TcHSC70-3 is a constitutively expressed protein that is also induced by heat. These results agreed with the reports that the HSP70 family includes the major heat shock proteins induced by thermal stresses [24, 26, 48]. Tchsp90 exhibited the same expression pattern as the two TcHSP70 genes, being significantly upregulated from 32 to 40°C. Similar response patterns have been reported in E. onukii, S. exigua and two Liriomyza species [21, 48, 53]. The upregulation of Tchsps expression in T. chilonis may contribute to the improvement of thermal tolerance. Some reports have documented that pupae of T. chilonis can endure a high temperature of 40°C [57, 58]. On the other hand, the heat-responsive expression of Tchsps also implies that the expression of Tchsps could be a potential indicator of the heat stress response [8, 16]. Further study is needed to support this speculation. Cold treatments (10 and 17°C) for 1 h led to a slight, but not significant, increase in the expression of all Tchsps, indicating that Tchsps are insensitive to low temperatures. In different insects, the expression patterns of these genes vary under cold exposure. For instance, hsp60 cannot be induced by cold in two Liriomyza species but could be upregulated in response to low temperature in C. chilonis [21, 24]. The expression of hsp70 decreases in response to cold shock in N. lugens while it increases in E. onukii [43, 53]. Some reports have indicated that the recovery from cold, rather than the direct cold stress, triggers the high expression of HSP genes [49, 59].
Under the 40°C treatment, the expression of all Tchsps exhibited a time-dependent response. Except for the expression level of Tchsp21.6 at 8 h, expression levels of these genes significantly increased after 1 h of exposure to 40°C and dramatically decreased at subsequent time points. This result indicated that the induction of hsps may be rapid and transitory. This phenomenon has also been reported in numerous studies [21, 31, 44]. Many studies have attributed the decrease in hsp expression to the energy balance and metabolic disorder [50]. In addition, previous studies have speculated that the synthesis of HSPs requires excess energy consumption, imposing stresses on various metabolic activities [60, 61]. The activity of enzymes is restricted under long-term heat exposure, which may also result in the decrease of hsp expression [62]. Under the 10°C treatment, the expression of Tchsp21.6, Tchsp60, Tchsc70-3 and Tchsp90 did not dramatically change at any time points. Tchsp10 and Tchsp70 exhibited a low-intensity cold response at 4 and 8 h. This result suggested that Tchsps have low or no sensitivity to cold temperatures.
Conclusions
In summary, six Tchsps were cloned and characterized from T. chilonis, namely, Tchsp10, Tchsp21.6, Tchsp60, Tchsp70, Tchsc70-3 and Tchsp90. These Tchsps exhibited different expression profiles at different developmental stages, suggesting they may be involved in the development of T. chilonis. In pupae of T. chilonis, the expression profiles of these genes could be induced by heat shocks (32 and 40°C for 1 h) but did not change in response to cold shocks (10 and 17°C for 1 h). In addition, their expression levels showed time-dependent responses to heat exposure. Tchsp10 and Tchsp70 exhibited a low-intensity cold response at 4 and 8 h. However, Tchsp21.6, Tchsp60, Tchsc70-3 and Tchsp90 did not respond to cold exposure. Due to the difficultly of sampling caused by the tiny size and parasitic characteristics of T. chilonis, our study initially explored the expression patterns of hsps during development and temperature stresses. Our study may aid in a better understanding of the roles of hsps at different development stages and in response to temperature stresses.
Supporting information
(DOCX)
Acknowledgments
We thank Ms. Xinxia Feng for the technical assistance and everyone who contributed to the development of this work at the Plant Protection Research Institute, Guangdong Academy of Agricultural Sciences.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was financially supported by the National Key R&D Plan (Grant No. 2017YFD0201000), the National Science Foundation for Young Scientists of China (Grant No. 31601631), the Fundamental Research Funds for the Central Universities (Grant No. 17lgpy109), the National Program on Key Basic Research Project (973 Program, Grant No. 2013CB127602). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sattar S, Farmanullah Saljoqi AR, Arif M, Sattar H, Qazi JI. Toxicity of some new insecticides against Trichogramma chilonis (Hymenoptera: Trichogrammatidae) under laboratory and extended laboratory conditions. Pak J Zool. 2011; 43(6): 1117–1125. [Google Scholar]
- 2.Zhang LH, Wang HS, Xu BF, Liu W. Classification and natural distribution of Trichogramma spp. in China. J Jilin Fores Sci Tech. 2014; 43(4), 607 33–35+38. [Google Scholar]
- 3.de Oliveira CM, de Oliveira JV, Barbosa DRES, Breda MO, de Franca SM, Duarte BLR. Biological parameters and thermal requirements of Trichogramma pretiosum for the management of the tomato fruit borer (Lepidoptera: Crambidae) in tomatoes. Crop Protection. 2017; 99: 39–44. 10.1016/j.cropro.2017.04.005 [DOI] [Google Scholar]
- 4.Nadeem S, Hamed M. Comparative development and parasitization of Trichogramma chilonis Ishii and Trichogrammatoidea bactrae Nagaraja under different temperature conditions. Pak J Zool. 2008; 40(6): 431–434. [Google Scholar]
- 5.Smith SM. Biological control with Trichogramma: advances, successes, and potential of their use. Annu Rev Entomol. 1996; 41: 375–406. 10.1146/annurev.en.41.010196.002111 [DOI] [PubMed] [Google Scholar]
- 6.Wang ZY, He KL, Zhang F, Lu X, Babendreier D. Mass rearing and release of Trichogramma for biological control of insect pests of corn in China. Biol Control. 2014; 68: 136–144. 10.1016/j.biocontrol.2013.06.015 [DOI] [Google Scholar]
- 7.Yi DW, Xiao R, Zhao YL, Li DS, Zhang GR. Cold storage of Corcyra cephalonica eggs affects the quality of Trichogramma chilonis offspring. J Environ Entomol. 2014; 36(4): 565–571. [Google Scholar]
- 8.Colinet H, Lee SF, Hoffmann A. Temporal expression of heat shock genes during cold stress and recovery from chill coma in adult Drosophila melanogaster. FEBS J. 2010; 277(1): 174–185. 10.1111/j.1742-4658.2009.07470.x [DOI] [PubMed] [Google Scholar]
- 9.Nadeem S, Ashfaq M, Hamed M, Ahmed S, Nadeem MK. Comparative rearing of Trichogramma chilonis (Ishii) (Hymenoptera: Trichogrammatidae) at different temperature conditions. Pak Entomol. 2009; 31(1): 33–36. [Google Scholar]
- 10.Kot J. Analysis of factors affecting the phytophage reduction by Trichogramma Westw. species. Pol Ecol Stud. 1979; 5: 5–59. [Google Scholar]
- 11.Hussain A, Razaq M, Saeed R, Aslam M, Rafiq M, Zaka SM. Effect of different temperatures on life history of Trichogramma chilonis (Ishii) in the laboratory conditions. Pak Entomol. 2013; 35(2): 123–127. [Google Scholar]
- 12.Tian JC, Wang ZC, Wang GR, Zhong LQ, Zheng XS, Xu HX, et al. The effects of temperature and host age on the fecundity of four Trichogramma species, egg parasitoids of the Cnaphalocrocis medinalis (Lepidoptera: Pyralidae). J Econ Entomol. 2017; 110(3): 949–953. 10.1093/jee/tox108 [DOI] [PubMed] [Google Scholar]
- 13.Liu QN, Zhu BJ, Dai LS, Fu WW, Lin KZ, Liu CL. Overexpression of small heat shock protein 21 protects the Chinese oak silkworm Antheraea pernyi against thermal stress. J Insect Physiol. 2013; 59(8): 848–854. 10.1016/j.jinsphys.2013.06.001 [DOI] [PubMed] [Google Scholar]
- 14.Chen W, Li D, Zhang M, Zhao Y, Wu W, Zhang G. Cloning and differential expression of five heat shock protein genes associated with thermal stress and development in the polyphagous predatory mite Neoseiulus cucumeris (Acari: Phytoseiidae). Exp Appl Acarol. 2015; 67(1): 65–85. 10.1007/s10493-015-9933-0 [DOI] [PubMed] [Google Scholar]
- 15.King AM, MacRae TH. Insect heat shock proteins during stress and diapause. Annu Rev Entomol. 2015; 60: 59–75. 10.1146/annurev-ento-011613-162107 [DOI] [PubMed] [Google Scholar]
- 16.Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999; 61: 243–282. 10.1146/annurev.physiol.61.1.243 [DOI] [PubMed] [Google Scholar]
- 17.Shen Y, Gu J, Huang LH, Zheng SC, Liu L, Xu WH, et al. Cloning and expression analysis of six small heat shock protein genes in the common cutworm, Spodoptera litura. J Insect Physiol. 2011; 57(7): 908–914. 10.1016/j.jinsphys.2011.03.026 [DOI] [PubMed] [Google Scholar]
- 18.Min Q, Cheng SY, Xi JF, Ma J, Xin TR, Xia B, et al. Expression patterns of three genes under short and long term cold exposure in Thitarodes pui (Lepidoptera: Hepialidae), a host of Ophiocordyceps sinensis. CryoLetters. 2016; 37(6): 432–439. [PubMed] [Google Scholar]
- 19.Yocum GD. Differential expression of two HSP70 transcripts in response to cold shock, thermoperiod, and adult diapause in the Colorado potato beetle. J Insect Physiol. 2001; 47(10): 1139–1145. [DOI] [PubMed] [Google Scholar]
- 20.Sonoda S, Fukumoto K, Izumi Y, Yoshida H, Tsumuki H. Cloning of heat shock protein genes (hsp90 and hsc70) and their expression during larval diapause and cold tolerance acquisition in the rice stem borer, Chilo suppressalis Walker. Arch Insect Biochem Physiol. 2006; 63(1): 36–47. 10.1002/arch.20138 [DOI] [PubMed] [Google Scholar]
- 21.Huang LH, Kang L. Cloning and interspecific altered expression of heat shock protein genes in two leafminer species in response to thermal stress. Insect Mol Biol. 2007; 16(4): 491–500. 10.1111/j.1365-2583.2007.00744.x [DOI] [PubMed] [Google Scholar]
- 22.Zou Z, Sun Z, Li J, Zhang G. Molecular cloning and characterization of two heat shock proteins in Thitarodes pui (Lepidoptera: Hepialidae). CryoLetters. 2011; 32(3): 225–239. [PubMed] [Google Scholar]
- 23.Wang H, Li K, Zhu JY, Fang Q, Ye GY, Wang H, et al. Cloning and expression pattern of heat shock protein genes from the endoparasitoid wasp, Pteromalus puparum in response to environmental stresses. Arch Insect Biochem Physiol. 2012; 79(4–5): 247–263. 10.1002/arch.21013 [DOI] [PubMed] [Google Scholar]
- 24.Pan D, Cao S, Lu M, Hang S, Du Y. Genes encoding heat shock proteins in the endoparasitoid wasp, Cotesia chilonis, and their expression in response to temperatures. J Integr Agr. 2018; 17(5) 1012–1022. 10.1016/s2095-3119(17)61737-4 [DOI] [Google Scholar]
- 25.Reineke A. Identification and expression of a small heat shock protein in two lines of the endoparasitic wasp Venturia canescens. Comp Biochem Physiol A Mol Integr Physiol. 2005; 141(1): 60–69. 10.1016/j.cbpb.2005.04.001 [DOI] [PubMed] [Google Scholar]
- 26.Xu P, Xiao J, Liu L, Li T, Huang D. Molecular cloning and characterization of four heat shock protein genes from Macrocentrus cingulum (Hymenoptera: Braconidae). Mol Biol Rep. 2010; 37(5): 2265–2272. 10.1007/s11033-009-9715-z [DOI] [PubMed] [Google Scholar]
- 27.Shi M, Wang YN, Zhu N, Chen XX. Four heat shock protein genes of the endoparasitoid wasp, Cotesia vestalis, and their transcriptional profiles in relation to developmental stages and temperature. PLoS ONE. 2013; 8(3): e59721 10.1371/journal.pone.0059721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volkoff AN, Daumal J, Barry P, Francois MC, Hawlitzky N, Rossi MM. Development of Trichogramma cacoeciae Marchal (Hymenoptera: Trichogrammatidae): time table and evidence for a single larval instar. Int J Insect Morphol Embryol. 1995; 24(4): 459–466. 10.1016/0020-7322(95)00009-s [DOI] [Google Scholar]
- 29.Huang YC, Yi DW, Song ZW, Li DS, Zhang GR. The individual development of Trichogramma chilonis on Corcyra cephalonica eggs. J Environ Entomol. 2016; 38 (3): 457–462. [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001; 25(4): 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 31.Xu D, Sun L, Liu S, Zhang L, Ru X, Zhao Y, et al. Molecular cloning of heat shock protein 10 (Hsp10) and 60 (Hsp60) cDNAs and their expression analysis under thermal stress in the sea cucumber Apostichopus japonicus. Comp Biochem Physiol B Biochem Mol Biol. 2014; 171: 49–57. 10.1016/j.cbpb.2014.03.009 [DOI] [PubMed] [Google Scholar]
- 32.Zhang B, Zheng J, Peng Y, Liu X, Hoffmann AA, Ma CS. Stress responses of small heat shock protein genes in Lepidoptera point to limited conservation of function across phylogeny. PLoS ONE. 2015; 10(7): e0132700 10.1371/journal.pone.0132700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi J, Fu M, Zhao C, Zhou F, Yang Q, Qiu L. Characterization and function analysis of Hsp60 and Hsp10 under different acute stresses in black tiger shrimp, Penaeus monodon. Cell Stress Chaperones. 2016; 21(2): 295–312. 10.1007/s12192-015-0660-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL–GroES–(ADP) 7 chaperonin complex. Nature. 1997; 388(6644): 741–750. 10.1038/41944 [DOI] [PubMed] [Google Scholar]
- 35.Hayer-Hartl M, Bracher A, Hartl FU. The GroEL–GroES chaperonin machine: a nano-cage for protein folding. Trends Biochem Sci. 2016; 41(1): 62–76. 10.1016/j.tibs.2015.07.009 [DOI] [PubMed] [Google Scholar]
- 36.Basha E, O’Neill H, Vierling E. Small heat shock proteins and α-crystallins: dynamic proteins with flexible functions. Trends Biochem Sci. 2012; 37(3): 106–117. 10.1016/j.tibs.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu QN, Liu Y, Xin ZZ, Zhu XY, Ge BM, Li CF, et al. A small heat shock protein 21 (sHSP21) mediates immune responses in Chinese oak silkworm Antheraea pernyi. Int J Biol Macromol. 2018; 111: 1027–1031. 10.1016/j.ijbiomac.2018.01.147 [DOI] [PubMed] [Google Scholar]
- 38.Franck E, Madsen O, van Rheede T, Ricard G, Huynen MA, de Jong WW. Evolutionary diversity of vertebrate small heat shock proteins. J Mol Evol. 2004; 59(6): 792–805. 10.1007/s00239-004-0013-z [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Zhao Q, Duan X, Song C, Chen M. Transcription of four Rhopalosiphum padi (L.) heat shock protein genes and their responses to heat stress and insecticide exposure. Comp Biochem Physiol A Mol Integr Physiol. 2017; 205: 48–57. 10.1016/j.cbpa.2016.12.021 [DOI] [PubMed] [Google Scholar]
- 40.Tang YC, Chang HC, Roeben A, Wischnewski D, Wischnewski N, Kerner MJ, et al. Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell. 2006; 125(5): 903–914. 10.1016/j.cell.2006.04.027 [DOI] [PubMed] [Google Scholar]
- 41.Singh MK, Reddy PVJ, Sreedhar AS, Tiwari PK. Molecular characterization and expression analysis of hsp60 gene homologue of sheep blowfly, Lucilia cuprina. J Therm Biol. 2015; 52: 24–37. 10.1016/j.jtherbio.2015.05.001 [DOI] [PubMed] [Google Scholar]
- 42.Wong CS, Mak CH, Ko RC. Cloning and characterization of the mitochondrial heat-shock protein 60 gene of Trichinella spiralis. Parasitol Res. 2004; 93(6): 461–467. 10.1007/s00436-004-1156-y [DOI] [PubMed] [Google Scholar]
- 43.Lu K, Chen X, Liu W, Zhang Z, Wang Y, You K. Characterization of heat shock protein 70 transcript from Nilaparvata lugens (Stål): Its response to temperature and insecticide stresses. Pestic Biochem Physiol. 2017; 142: 102–110. 10.1016/j.pestbp.2017.01.011 [DOI] [PubMed] [Google Scholar]
- 44.Cheng W, Li D, Wang Y, Liu Y, Zhu-Salzman K. Cloning of heat shock protein genes (hsp70, hsc70 and hsp90) and their expression in response to larval diapause and thermal stress in the wheat blossom midge, Sitodiplosis mosellana. J Insect Physiol. 2016; 95: 66–77. 10.1016/j.jinsphys.2016.09.005 [DOI] [PubMed] [Google Scholar]
- 45.Chen HL, Zhang HY, Throne JE, Zhu KY. Transcript analysis and expression profiling of three heat shock protein 70 genes in the ectoparasitoid Habrobracon hebetor (Hymenoptera: Braconidae). Insect Sci. 2014; 21(4): 415–428. 10.1111/1744-7917.12032 [DOI] [PubMed] [Google Scholar]
- 46.Ju RT, Luo QQ, Gao L, Yang J, Li B. Identification of HSP70 gene in Corythucha ciliata and its expression profiles under laboratory and field thermal conditions. Cell Stress Chaperones. 2018; 23(2): 195–201. 10.1007/s12192-017-0840-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang LH, Wang CZ, Kang L. Cloning and expression of five heat shock protein genes in relation to cold hardening and development in the leafminer, Liriomyza sativa. J Insect Physiol. 2009; 55(3): 279–285. 10.1016/j.jinsphys.2008.12.004 [DOI] [PubMed] [Google Scholar]
- 48.Jiang X, Zhai H, Wang L, Luo L, Sappington TW, Zhang L. Cloning of the heat shock protein 90 and 70 genes from the beet armyworm, Spodoptera exigua, and expression characteristics in relation to thermal stress and development. Cell Stress Chaperones. 2012; 17(1): 67–80. 10.1007/s12192-011-0286-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H, Reitz SR, Wang L, Wang S, Li X, Lei Z. The mRNA expression profiles of five heat shock protein genes from Frankliniella occidentalis at different stages and their responses to temperatures and insecticides. J Integr Agr. 2014; 13(10): 2196–2210. 10.1016/S2095-3119(13)60680-2 [DOI] [Google Scholar]
- 50.Tan Y, Zhang Y, Huo ZJ, Zhou XR, Pang BP. Molecular cloning of heat shock protein 10 (Hsp10) and 60 (Hsp60) cDNAs from Galeruca daurica (Coleoptera: Chrysomelidae) and their expression analysis. Bull Entomol Res. 2017; 1–13. 10.1017/S0007485317001079 [DOI] [PubMed] [Google Scholar]
- 51.Zhao L, Jones WA. Expression of heat shock protein genes in insect stress responses. Isj-Invert Surviv J. 2012; 9: 93–101. [Google Scholar]
- 52.Sørensen JG, Kristensen TN, Loeschcke V. The evolutionary and ecological role of heat shock proteins. Ecol Lett. 2003; 6(11): 1025–1037. 10.1046/j.1461-0248.2003.00528.x [DOI] [Google Scholar]
- 53.Qiao L, Wu JX, Qin DZ, Liu XC, Lu ZC, Lv LZ, et al. Gene expression profiles of heat shock proteins 70 and 90 from Empoasca onukii (Hemiptera: Cicadellidae) in response to temperature stress. J Insect Sci. 2015; 15(1): 49 10.1093/jisesa/iev030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tazir Y, Steisslinger V, Soblik H, Younis AE, Beckmann S, Grevelding CG, et al. Molecular and functional characterization of the heat shock protein 10 of Strongyloides ratti. Mol Biochem Parasitol. 2009; 168(2): 149–157. 10.1016/j.molbiopara.2009.07.007 [DOI] [PubMed] [Google Scholar]
- 55.Liu WW, Yang P, Chen XM, Xu DL, Hu YH. Cloning and Expression Analysis of Four Heat Shock Protein Genes in Ericerus pela (Homoptera: Coccidae). J Insect Sci. 2014; 14(1): 142 10.1093/jisesa/ieu032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang H, Fang Y, Wang L, Zhu W, Ji H, Wang H, et al. Transcriptome analysis of the Bombyx mori fat body after constant high temperature treatment shows differences between the sexes. Mol Biol Rep. 2014; 41(9): 6039–6049. 10.1007/s11033-014-3481-2 [DOI] [PubMed] [Google Scholar]
- 57.Firake DM, Khan MA. Alternating temperatures affect the performance of Trichogramma species. J Insect Sci. 2014; 14(1): 41–41. 10.1093/jis/14.1.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dadmal SM, Pujari AJ, Satpute NS. Influence of short term exposure to different temperatures on key biological parameters of Trichogramma chilonis Ishii under laboratory conditions. J Biol Control. 2010; 24(1): 8–12. [Google Scholar]
- 59.Sinclair BJ, Gibbs AG, Roberts SP. Gene transcription during exposure to, and recovery from, cold and desiccation stress in Drosophila melanogaster. Insect Mol Biol. 2007; 16(4): 435–443. 10.1111/j.1365-2583.2007.00739.x [DOI] [PubMed] [Google Scholar]
- 60.Krebs RA, Feder ME. Deleterious consequences of Hsp70 overexpression in Drosphilla melanogaster larvae. Cell Stress Chaperones. 1997; 2(1): 60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krebs RA, Feder ME. Hsp70 and larval thermotolerance in Drosophila melanogaster: how much is enough and when is more too much? J Insect Physiol. 1998; 44(11): 1091–1101. [DOI] [PubMed] [Google Scholar]
- 62.Sokolova I, Pörtner HO. Temperature effects on key metabolic enzymes in Littorina saxatilis and L. obtusata from different latitudes and shore levels. Mar Biol. 2001; 139(1): 113–126. 10.1007/s002270100557 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.