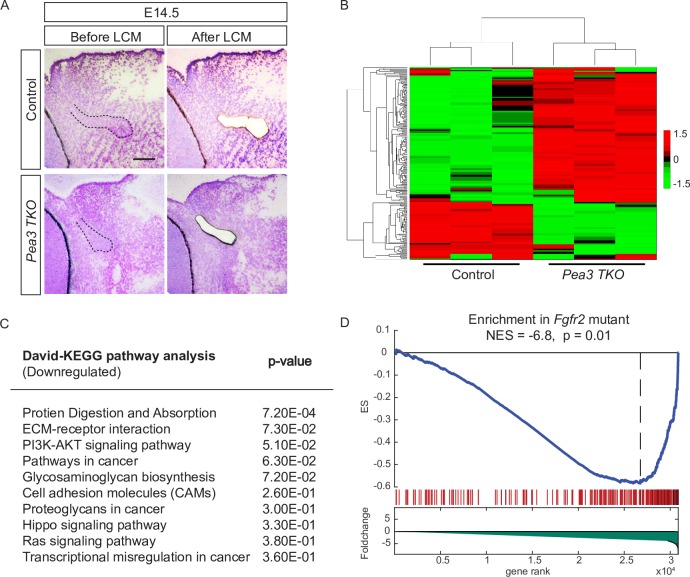
Fig 2. Bioinformatics analysis indicates that Pea3 transcription factors are downstream of FGF signaling during lacrimal gland development.
(A) Images of sections before and after laser capture microscopy to isolate the lacrimal gland bud. Scale bar, 50μm. (B) Clustergram analysis of the top 200 differentially expressed genes in RNA-seq data from 6 different samples (Control, n = 3, Mutant, n = 3). The control and mutant samples were segregated into two separate clusters, indicating the robustness of our data (r = 0.8). (C) KEGG pathway analysis of downregulated genes in Pea3 mutant using DAVID indicated that several key pathways were significantly downregulated. (D) Gene set enrichment analysis (GSEA) analysis showed that the transcriptome changes in Pea3 mutants resemble that of the previously reported Fgfr2 mutant.