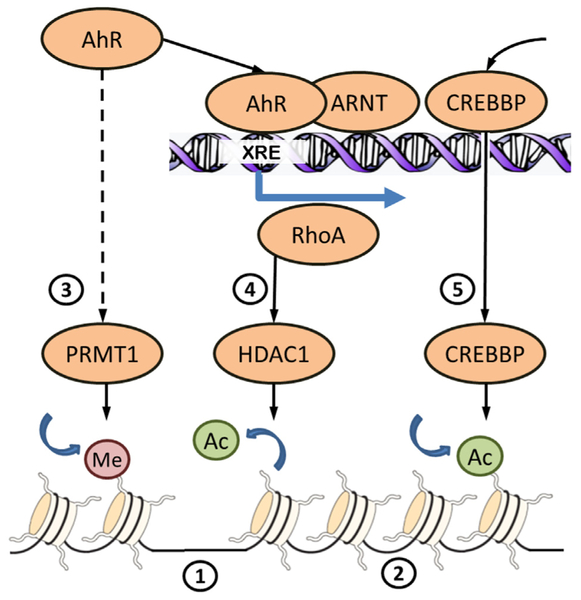
Fig. 2.
Some mechanisms of chromatin rearrangement downstream AhR signaling. DNA in somatic cells is wrapped around nucleosomes consisting of histone proteins. Sparse position of nucleosomes (1) insures accessibility of DNA for transcription machinery and is associated with highly expressed genes. DNA tightly packed on nucleosomes (2) is associated with inactive genes. Compaction and relaxation of chromatin is regulated by covalent modification of histone tails. Activation of AhR by ligand-binding activates histone methyltransferase PRMT1 (3) by unknown mechanisms. PRMT1 methylates arginine 3 of histone 4 what favors further acetylation of histone 4. Acetylation neutralizes the positive charges on the histone, decreases histone–DNA binding and results in transcriptional activation. Binding of AhR/ARNT heterodimer to xenobiotic response element (XRE) initiates transcription of genes, including RhoA, resulting in increased expression and nuclear translocation of histone deacetylase HDAC1 (4). Histone deacetylation condenses the chromatin structure resulting in the downregulation of target genes. HDAC1 participates in repression of cell cycle genes. Binding of AhR/ARNT to XRE recruits histone acetyltransferase CREBBP (5). Aacetylation of histone tails by CREBBP participates in transcription initiation