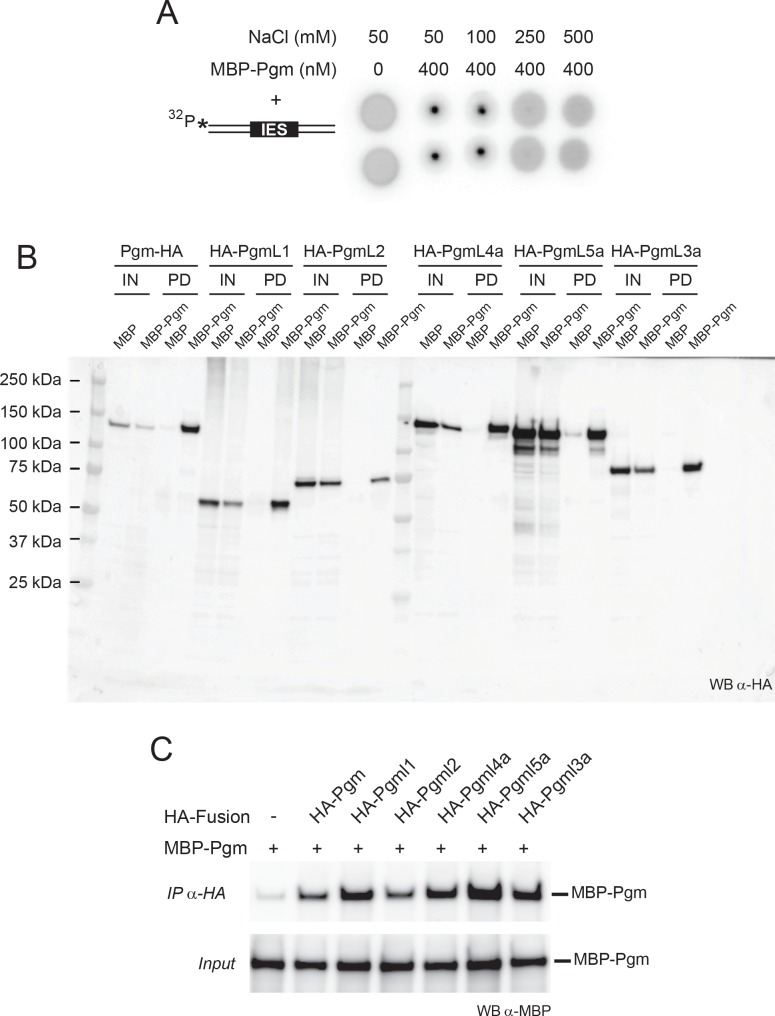
Figure 3. PgmLs are essential during autogamy and interact with Pgm in cell extracts.
(A) Effect of PGML KDs on the recovery of post-autogamous progeny with functional new MACs. For PGML1, PGML2 and PGML3c, only the results obtained using IF1 RNAi constructs (Figure 3—figure supplement 1) are shown. For groups of duplicated paralogs, individual gene KDs were performed using gene-specific IF2 constructs (Figure 3—figure supplement 1), while double KDs were performed using either IF2 or cross-hybridizing IF1 (*) constructs. Error bars represent standard deviations (n = 2 to 14, see Supplementary file 6) (B) Pull down of HA-PgmL fusions with MBP-Pgm using recombinant proteins expressed in insect cells. In each panel, the HA-tagged protein that was co-expressed with MBP or MBP-Pgm is indicated on the left and the band revealed on western blots (WB) using anti-HA antibodies is indicated on the right. The full-size blot with molecular weight marker is shown in Figure 3—figure supplement 2.
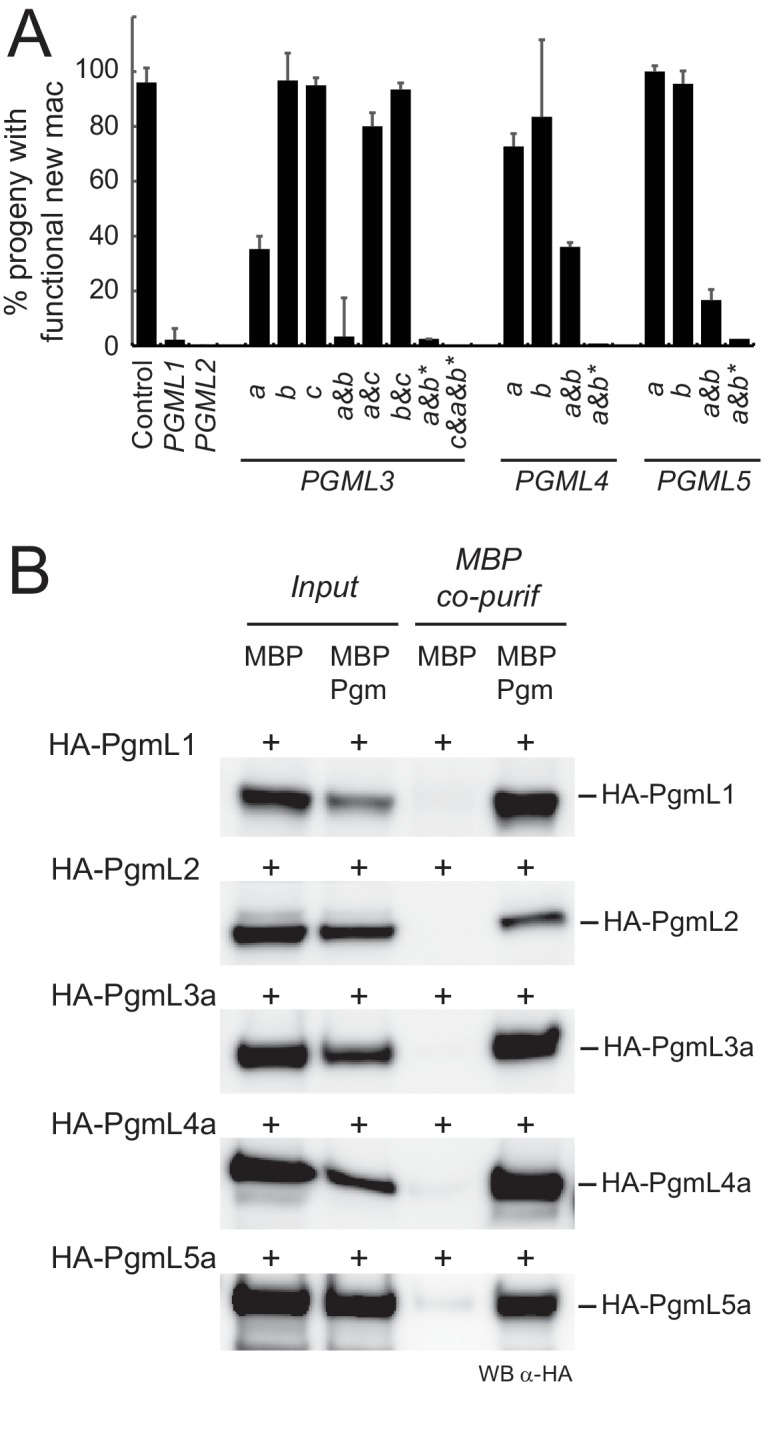
Figure 3—figure supplement 1. Map and coordinates of PGML feeding inserts.