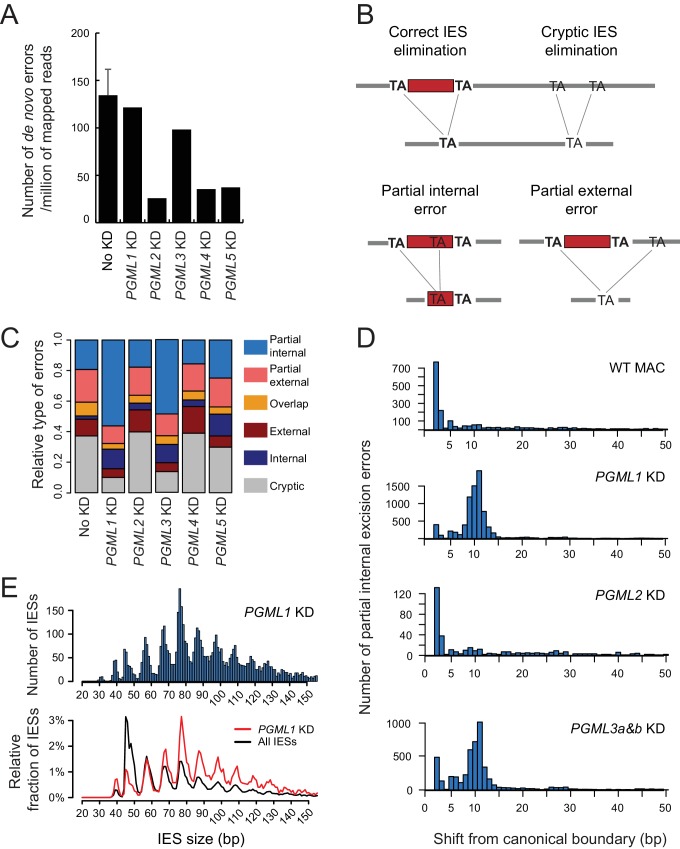
Figure 6. IES excision errors in PGML KDs.
(A) Number of IES excision errors in the MAC of vegetative cells before autogamy (No KD) and in the new MAC of autogamous cells upon each PGML KD (de novo errors). For the No KD sample, the error bar represents the standard deviation for five replicates (V samples in Supplementary file 9). (B) Major classes of IES excision errors found in the different samples. In external or internal errors, the two alternative TAs are misplaced (one on each flanking side or both inside of a reference IES, respectively). Overlapping errors use one TA inside and the other outside of reference IESs. (C) Distribution of the different classes of de novo excision errors in PGML KDs. As a control, the distribution of pre-existing errors found in the old MAC is shown for a vegetative culture (see Supplementary file 9). (D) Position of alternative excision boundaries used in partial internal excision errors, relative to the canonical boundary of the reference IES. WT: vegetative MAC; for PGML KDs, only de novo errors were considered. (E) Size distribution of IESs exhibiting partial internal errors in a PGML1 KD. Upper panel: size distribution of all IESs with partial internal errors. Lower panel: the black curve shows the fraction of IESs of each size relative to the total number of IESs in the genome; the red curve shows the fraction of IESs of each size among the population of IESs showing at least one partial internal error in a PGML1 KD. In both panels, only IESs with an alternative boundary at >2 bp from the canonical one were counted. In the bottom panel, IESs shorter than 35 bp were not considered (see Figure 6—figure supplement 4).
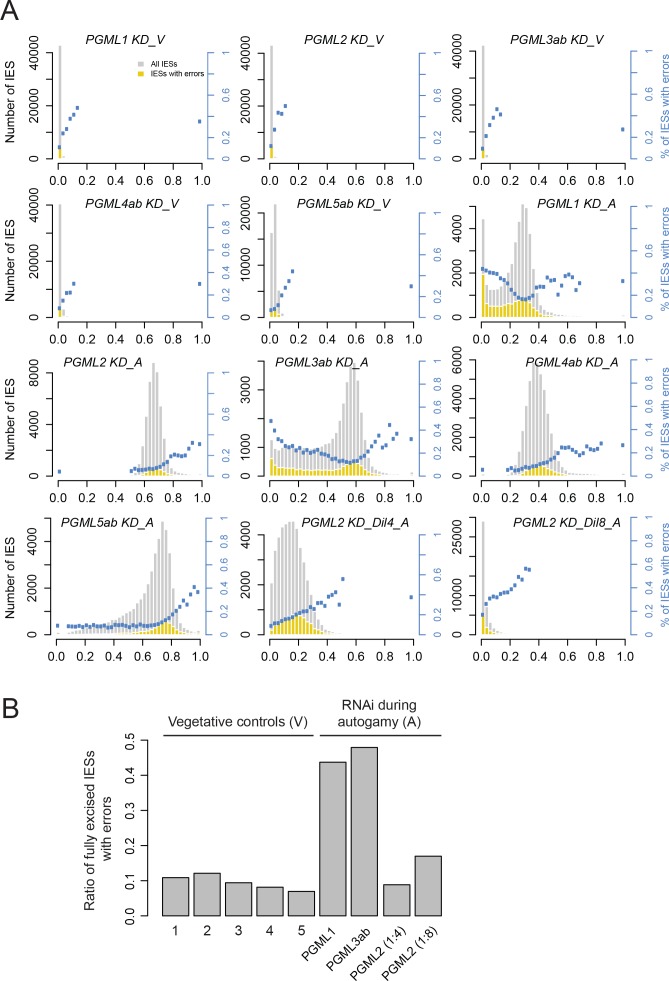
Figure 6—figure supplement 1. The number of excision errors increases for IESs with the lowest retention scores in PGML1 or PGML3a and b knockdowns.
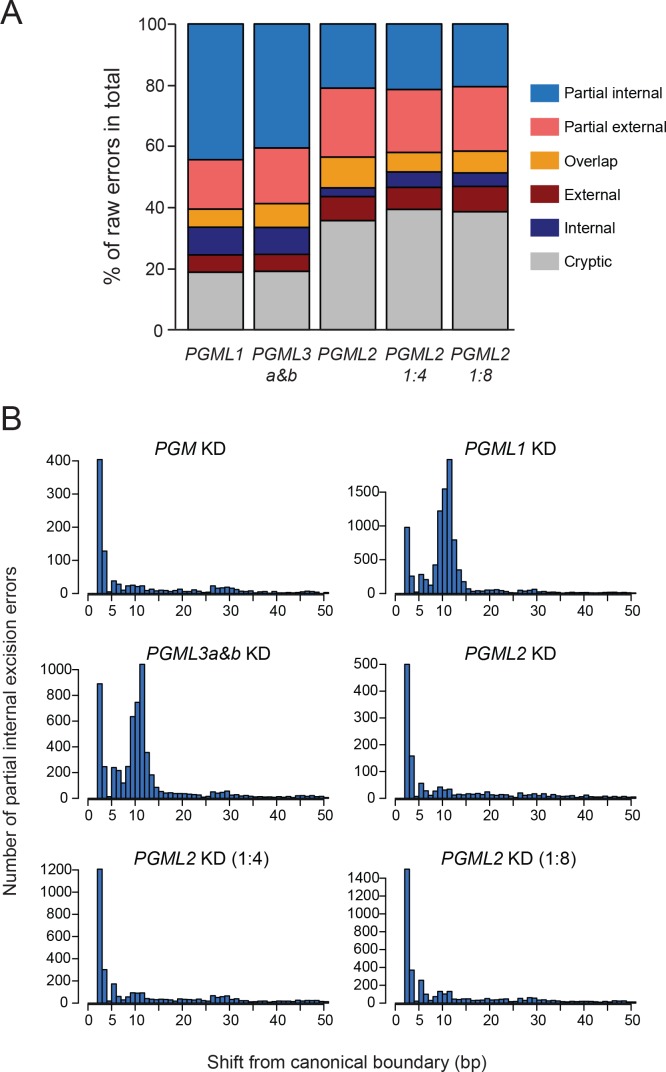
Figure 6—figure supplement 2. Raw counts of IES excision errors in PGML1, PGML3a and b and partial PGML2 knockdowns.
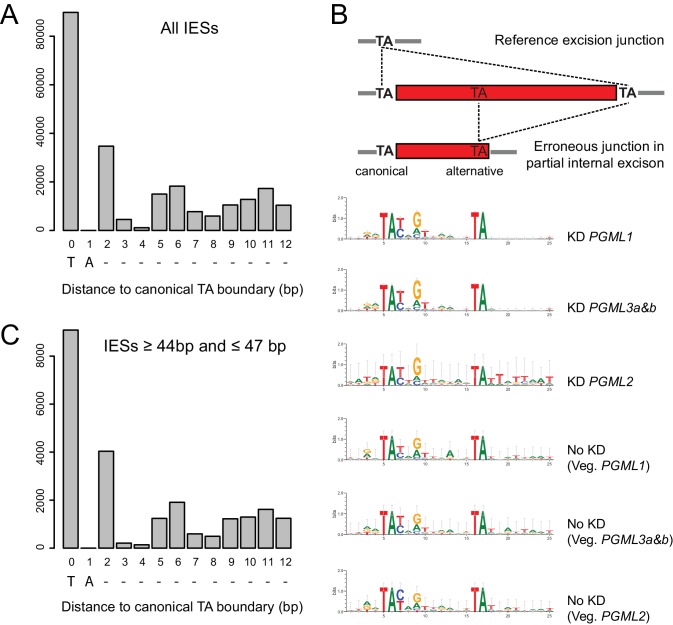
Figure 6—figure supplement 3. Alternative excision boundaries used in partial internal IES excision errors.
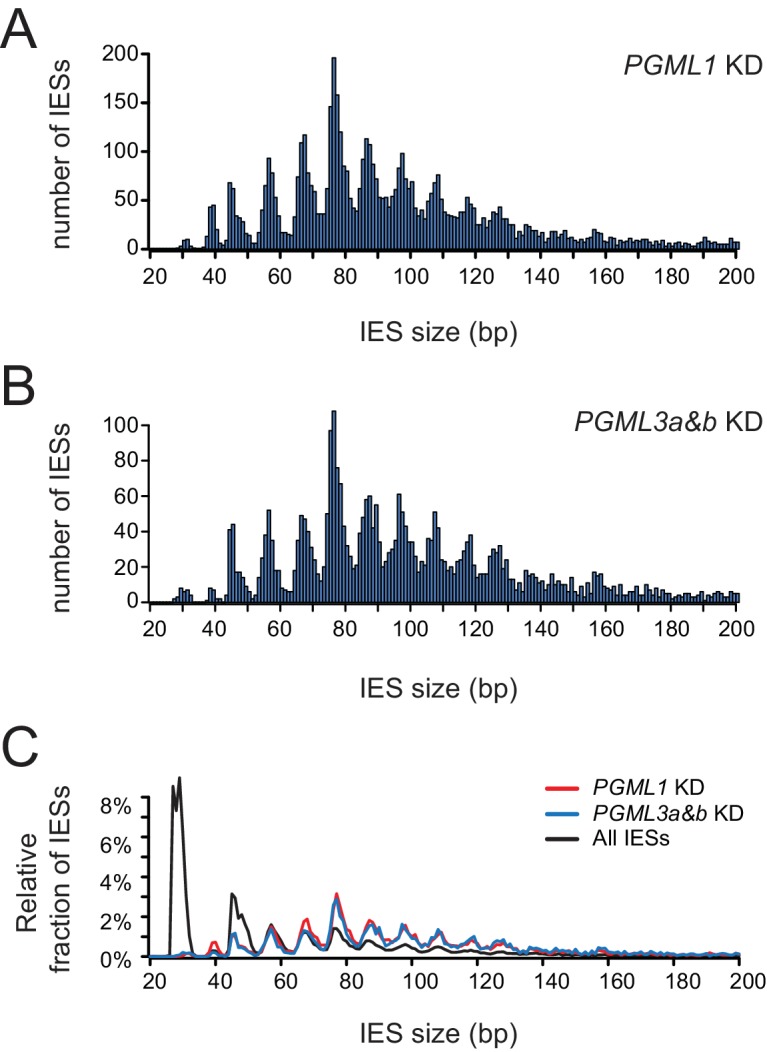
Figure 6—figure supplement 4. Size distribution of IESs with partial internal excision errors in PGML1 or PGML3a and b KDs.