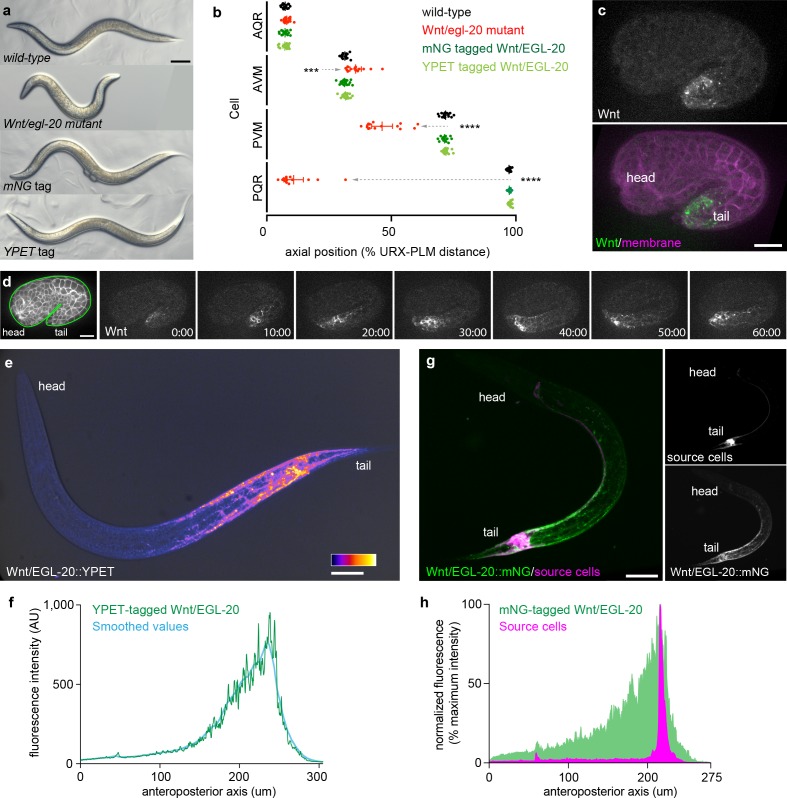
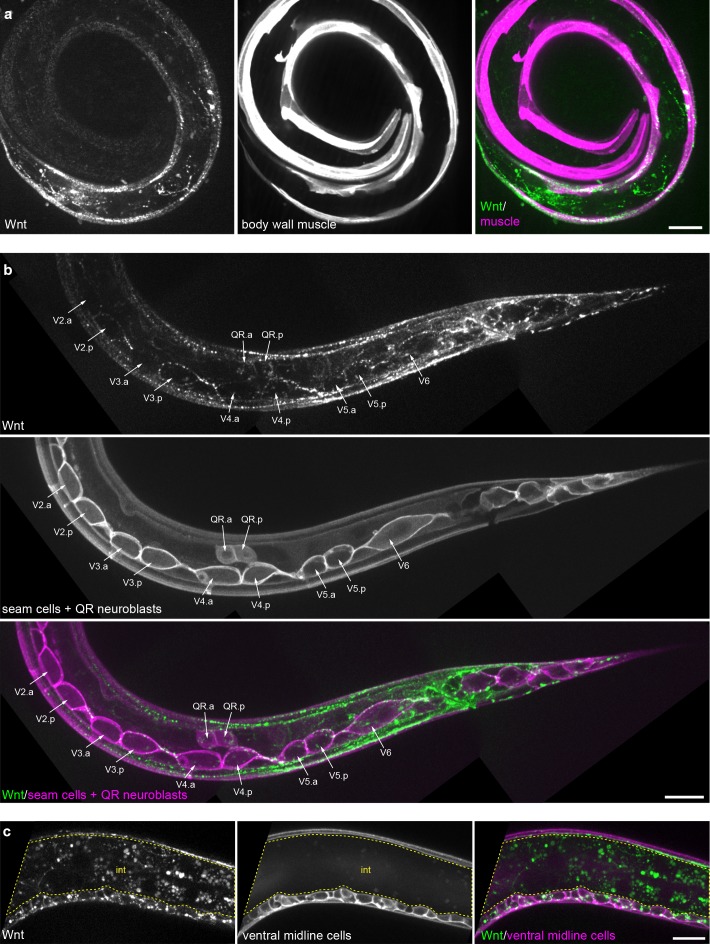
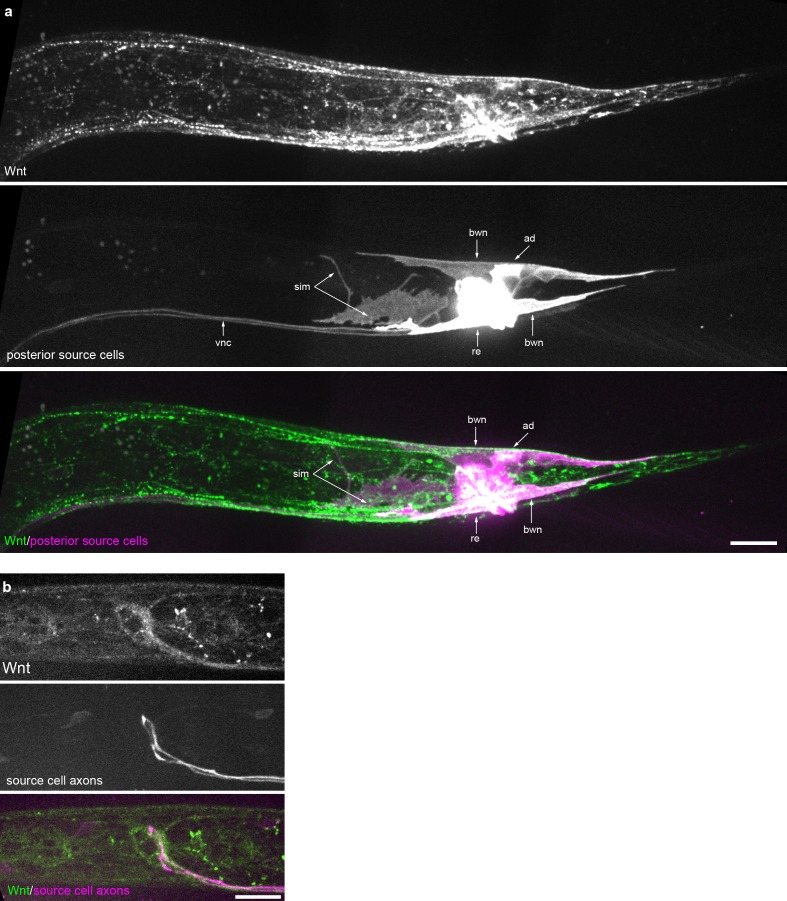
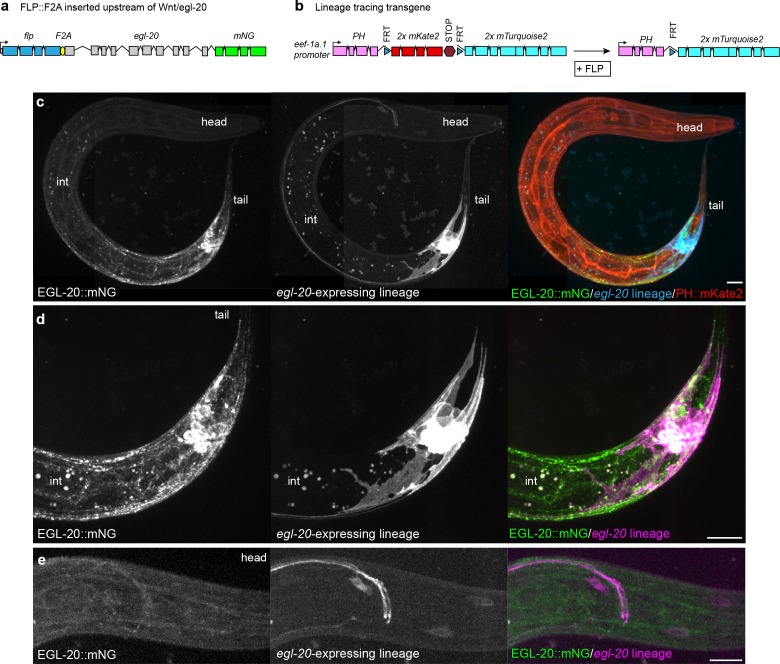
Figure 1. Tagged Wnt/EGL-20 is biologically functional and forms a long-range, anteroposterior gradient in vivo.
(a) transmitted light images of adult C. elegans with wild-type egl-20, the egl-20 loss-of-function mutant egl-20(n585), mNG-tagged egl-20, or YPET-tagged egl-20 showing normal external anatomy in mNG and YPET-tagged strains; (b) positions of QR neuroblast descendants AQR and AVM and QL neuroblast descendants PVM and PQR in wild-type, egl-20 mutant, egl-20::mNG, and egl-20::YPET strains showing that tagged EGL-20 is biologically functional for Q neuroblast migration. Dashed arrows indicate abnormal cell migrations. Means and 95% confidence intervals are shown for each cell type/genotype. Wild-type n = 15, egl-20(n585) n = 15, EGL-20::mNG n = 20, EGL-20::YPET n = 18.***, adjusted p=0.0005; ****, adjusted p<0.0001, all other comparisons adjusted p>0.9999, one-way ANOVA with Sidak’s multiple comparisons test; (c) maximum intensity projection of a comma stage embryo showing the earliest detectable Wnt/EGL-20::mNG fluorescence; (d) surface optical sections from time-lapse images of Wnt/EGL-20::mNG showing the onset of spreading from 1.5-fold to 2-fold stages; (e) maximum intensity projection of Wnt/EGL-20::YPET fluorescence in a living, late L1 stage animal illustrating the anteroposterior Wnt gradient colored with fire look-up-table and overlaid with transmitted light image; (f) profile plot of raw and LOWESS smoothed Wnt/EGL-20::YPET fluorescence intensity along the anteroposterior axis in the same worm as in (e); (g) maximum intensity projections of a living, mid L1 stage animal showing plasma membranes of egl-20 source cells labeled by Pegl-20>2x mKate2::PH (magenta) and Wnt/EGL-20::mNG protein (green); (h) profile plot of normalized EGL-20::mNG and Pegl-20>2x mKate2::PH fluorescence intensities along the anteroposterior axis illustrating Wnt dispersal from source cells. Images are oriented with anterior to left and dorsal to top. Scale bars = 0.1 mm in a, 10 μm in c and d, and 20 μm in e and g.