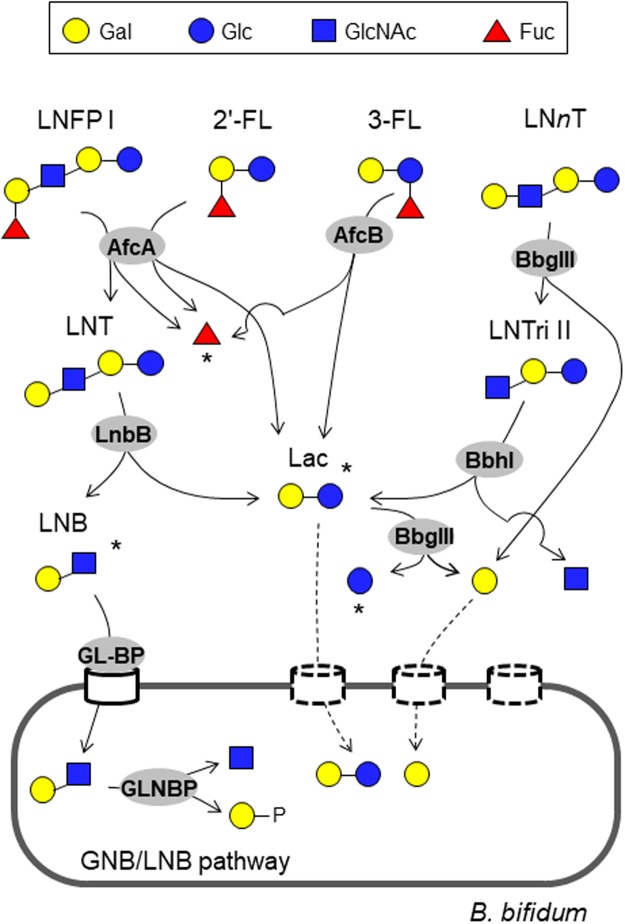
Figure 1.
Schematic representation of the minimum enzymatic set required to degrade different sugar linkages found in neutral HMOs by B. bifidum (See also Table 1). Sugars are depicted according to the nomenclature committee of the Consortium for Function Glycomics. LNFP I (Fucα1-2Galβ1-3GlcNAcβ1-3Galβ1-4Glc), 2′-FL (Fucα1-2Galβ1-4Glc), 3-FL (Galβ1-4(Fucα1-3)Glc), and LNnT (Galβ1-4GlcNAcβ1-3Galβ1-4Glc) are shown as representatives of the different sugar linkages. AfcA: 1,2-α-l-fucosidase; AfcB: 1,3-1,4-α-l-fucosidase; LnbB: lacto-N-biosiadse; BbgIII: β-1,4-galactosidase; BbhI: β-N-acetylglucosaminidase; GL-BP: galacto-N-biose/lacto-N-biose I-binding protein of ABC transporter; GLNBP: galacto-N-biose/lacto-N-biose I phosphorylase. Dashed lines indicate pathways that have not been identified in B. bifidum. Degradants that accumulate in the spent media during culture in the presence of HMOs are indicated by asterisks (See Fig. 3 and Table S3).