Fig. 5.
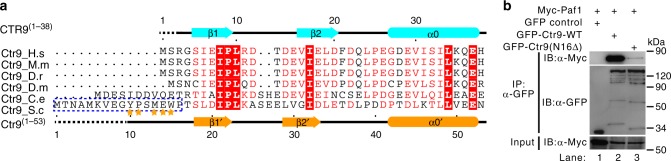
The longer N-terminal tail of Ctr9 is essential for its binding to Paf1. a Structural-based sequence alignment of the N-terminal fragments of Ctr9 in various species. In this alignment, the secondary structures of human CTR9(1–38) and yeast Ctr9(1–53) are shown at the top and bottom, respectively, and conserved residues are shaded in red. The amino acids 1–16 of yeast Ctr9, which were deleted in the GFP-Ctr9(N16Δ) construct [used in b], are marked with a dotted blue box. The amino acids Y10, P11, M13, E14, and W15 of Ctr9 involved in its binding to Paf1 are marked by orange stars. b Co-IP experiments testing the interactions between Ctr9 WT or Ctr9(N16Δ) mutant and Paf1. Extracts were prepared from HEK293T cells transfected with various combinations of plasmids, as indicated. The bottom panel shows 3% of the Myc fusion proteins for each IP. Uncropped blots are shown in Supplementary Fig. 8