FIGURE 7.
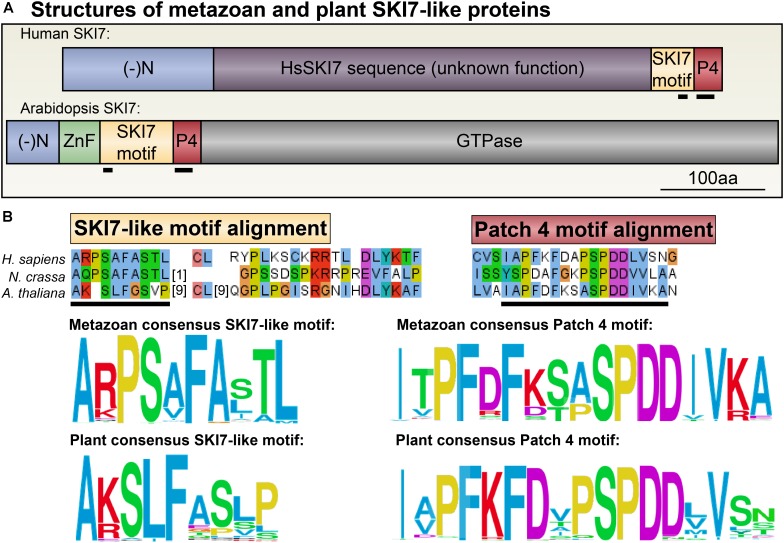
Consensus amino acid sequences that define eukaryotic SKI7. (A) Human and Arabidopsis SKI7 proteins do not globally align. Human SKI7 includes a long insert of unknown function (pink) between the negatively charged N-terminus (blue) and the SKI7- and Patch 4-like motifs (red). Arabidopsis SKI7 does not include this insert, but does include a C-terminal trGTPase domain of unclear function for SKI7, but which is required for HBS1 decoding activity. (B) Alignments of NCBI RefSeq SKI7 proteins from metazoans (Supplementary Data Sheet 2) and plants (Supplementary Data Sheet 1) were analyzed to determine consensus sequences for the SKI7-like motifs and the Patch 4-like motifs in these two major eukaryotic lineages. The sequences are largely similar, although the metazoan SKI7-like motif includes a widely conserved proline insertion (at position 3 in the consensus sequence). Alignments of the human, Neurospora crassa (fungus), and Arabidopsis SKI7-like motif and Patch 4-like motifs, as well as surrounding residues, are shown above the consensus sequences as examples. Amino acids are colored following Clustal standards, as above (Figure 5A).