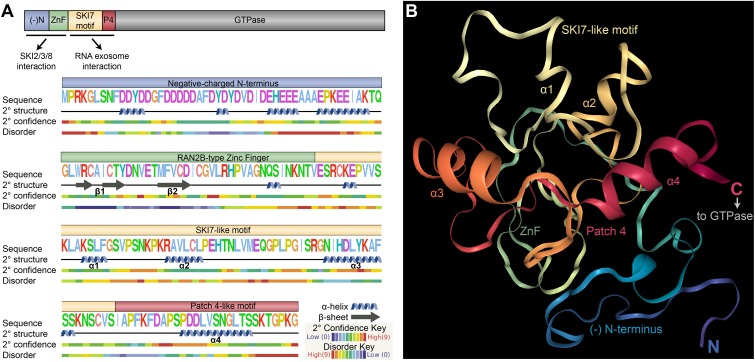
FIGURE 8.
Predicted structure of the Arabidopsis SKI7 N-terminus. (A) Phyre2 prediction of the secondary structure and disorder of the N-terminal 183 amino acids of Arabidopsis SKI7. The negatively charged N-terminus (blue, (-)N) and RAN2B-type Zinc Finger (green, ZnF) are predicted to interact with the SKI2/SKI3/SKI8 cytosolic exosome adaptor complex, and the SKI7-like motif (yellow) and Patch 4-like motif (red, P4) are predicted to interact with the surface of the RNA exosome catalytic core. The negatively charged N-terminus includes moderate-confidence α-helices that may promote interaction with SKI2/SKI3/SKI8. The ZnF domain includes two β-sheets (β1 and β2) and an α-helix that coordinate with the Zn ion. The SKI7-like and Patch 4-like motif include four α-helices (α1 through α4), which is structurally comparable to the yeast SKI7 protein. (B) Phyre2 prediction of the structure of the N-terminal 183 amino acids of Arabidopsis SKI7, based on homology modeling against several resolved protein structures (as described in the methods). Residues are colored by position, from blue (N-terminus) to red (C-terminus), closely matching the colors used in panel a and other figures. The defined regions of the protein are labeled, including α1, α2, α3, and α4, which are predicted to mediate interactions with the RNA exosome core. The C-terminus of this structure is predicted to be highly disordered, forming a flexible linker to the C-terminal trGTPase of Arabidopsis SKI7.