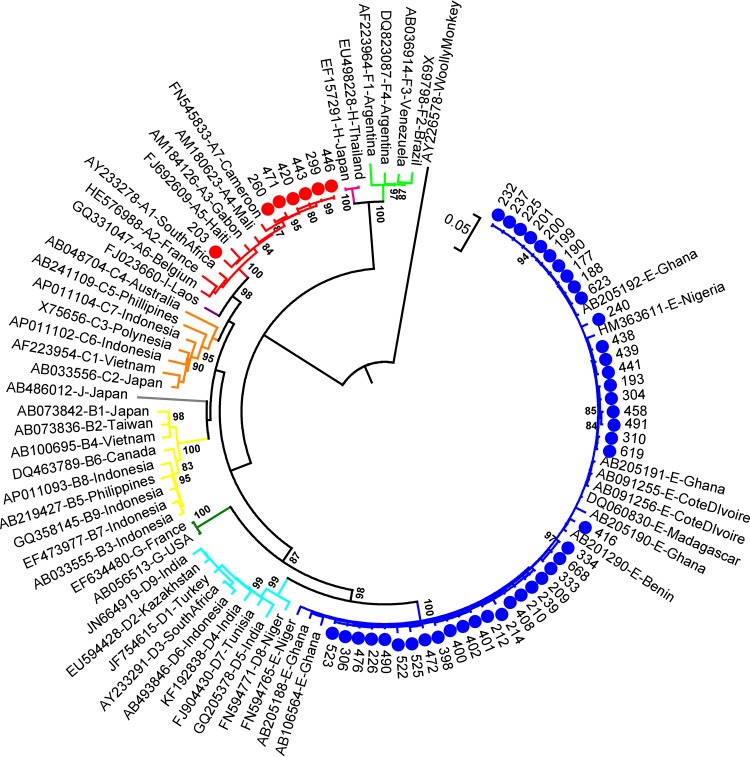
FIG 2.
Phylogenetic reconstruction of HBV pre-S/S sequences. A maximum-likelihood phylogenetic tree of the 49 HBV pre-S/S sequences obtained in this study together with 53 publicly available sequences covering all HBV genotypes was constructed under the general time-reversible (GTR) model +G +I embedded in MEGA 6.0. Of the 49 Mbandji 2 sequences, 42 (blue dots) and 7 (red dots) clustered with HBV/E and HBV/A isolates, respectively. The tree was drawn to scale, with branch lengths measured as the number of substitutions per site. The tree was rooted using the sequence of a woolly monkey HBV as an outgroup. There were a total of 1,212 positions in the final data set. Bootstrap values (≥80%) are shown at branch nodes.