Abstract
The aim of the present study was to measure the expression of Claudin (CLDN) 1 in nasopharyngeal carcinoma (NPC) and to determine its biological function and mechanism of action. Reverse transcription-quantitative polymerase chain reaction and western blotting were performed to measure the expression of CLDN1 mRNA and protein, respectively, in the immortalized human nasopharyngeal epithelial cell line NP69 and NPC-TW01 cells. Subsequently, small interfering RNA against CLDN1 and the LV-GFP-PURO-CLDN1 lentivirus were transfected into NPC-TW01 cells. Western blotting was used to determine the effects of CLDN1 down- and upregulation on the expression of the epithelial mesenchymal transition (EMT) markers E-cadherin and vimentin. In addition, the effect of CLDN1 on the expression of β-Catenin was determined. The results demonstrated that levels of CLDN1 mRNA and protein in NPC cells were significantly higher than in NP69 cells. Furthermore, the downregulation of CLDN1 inhibited the proliferation, invasion and migration of NPC-TW01 cells. The results of western blotting demonstrated that the downregulation of CLDN1 resulted in the upregulation of E-cadherin and inhibition of vimentin in NPC-TW01 cells. By contrast, the overexpression of CLDN1 resulted in the downregulation of E-cadherin and upregulation of vimentin in NPC-TW01 cells. The downregulation of β-catenin attenuated the cancer-promoting effect of CLDN1 on NPC-TW01 cells, whereas the upregulation of β-catenin reversed the tumor-suppressing effect of CLDN1 downregulation on NPC-TW01 cells. The results of the present study therefore demonstrate that CLDN1 expression is elevated in NPC cells. As an oncogene, CLDN1 promotes the proliferation, invasion and migration of NPC cells by upregulating the expression and nuclear entry of β-catenin.
Keywords: claudin-1, nasopharyngeal carcinoma, β-catenin
Introduction
Nasopharyngeal carcinoma (NPC) is a malignant tumor affecting the head and neck and has a particularly high incidence in Southern China (1,2). The onset and development of NPC is closely associated with genetic factors, Epstein-Barr viral infection, air pollution and excessive contact with trace elements, including nickel (3). Poorly differentiated squamous cell carcinoma is the most common type of NPC, whereas the incidence of other types of NPC, including highly differentiated squamous cell carcinoma, adenocarcinoma and vesicular nucleus cell carcinoma, is much lower (4). Most patients with NPC are sensitive to radiotherapy, which is the preferred modality of treatment (5).
The overall survival rate of patients with NPC has been increasing; however the prognosis of patients with advanced NPC remains poor and the 5-year survival rate of such patients is only 30–40% (6,7). Tumor recurrence and distant metastasis are the primary factors affecting the prognosis of patients with NPC (8). It has been demonstrated that 70–80% of patients with NPC already have lymph node metastasis upon diagnosis and 60% of patients with NPC experience neck lymph node enlargement as their initial symptom (9). Therefore, it is important to determine the molecular mechanism of invasion and metastasis of NPC, as this may improve the early diagnosis and treatment of patients with NPC.
Tight junctions are important structures that maintain cell connections and polarity and are composed of a variety of proteins (10). The disordered expression of tight junctions is closely associated with the proliferation, invasion, metastasis and apoptosis of tumors (11,12). Claudin (CLDN) 3 and CLDN4 are important proteins in the intestinal epithelial cell barrier and their disordered expression induces tumor metastasis (13,14). CLDN3 expression is downregulated in early gastric cancer, whereas CLDN4 overexpression is closely associated with the development of gastric cancer (15,16). Furthermore, CLDN6, CLDN7 and CLDN9 promote the proliferation, invasion and metastasis of gastric cancer cells (17,18).
CLDN1 is one of the key structural proteins in the tight junction protein family and is distributed across the surface of the cell membrane in the form of a transmembrane structure that crosses the membrane four times. CLDN1 interacts with the isoenzymes of creatine kinase, tight junction proteins ZO1, ZO2, ZO3 and proteins containing the PDZ domain, to pass signals inside and outside cells and maintain the physical barrier function of tight junctions (19,20). The expression of CLDN1 is upregulated in NPC and is associated with lymph node metastasis and clinical staging; however, the function of CLDN1 in NPC remains unclear (21).
The epithelial-mesenchymal transition (EMT) is a process during which epithelial cells lose their polarity and gain interstitial cell properties, including increased invasiveness and metastasis, and decreased rates of apoptosis. The EMT is a key step in tumor metastasis and inhibiting the EMT thus inhibits tumor invasion and metastasis (22). Previous studies have demonstrated that numerous genes and signaling pathways are involved in the EMT in epithelial tumors. Inhibiting enhancer of zeste homolog 2 expression and enhancing forkhead box protein C2 expression promotes the EMT in ovarian cancer cells (23). In addition, microRNA-630 regulates the invasion and metastasis of esophageal squamous carcinoma cells by inhibiting the EMT (24). Tight junction proteins are associated with the EMT in tumors (25); however, to the best of our knowledge, the effect of CLDN1 on the EMT in NPC cells has not yet been reported. It has also been demonstrated that the Wnt/β-catenin signaling pathway serves an important role in the EMT and β-catenin aggregation in cytoplasm stimulates the entry of β-catenin into the nucleus. This triggers the transcription of EMT-related genes and the onset of the EMT (26). The present study assessed the biological effects and molecular mechanisms of CLDN1 in NPC, with the aim of determining the effect of tight junction proteins in NPC.
Materials and methods
Cells
The human nasopharyngeal epithelial cell line NP69 was purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). The human nasopharyngeal carcinoma cell line NPC-TW01 was purchased from the American Type Culture Collection (Manassas, VA, USA). All cells were cultured in RPMI-1640 medium containing 10% fetal bovine serum (both obtained from BD Biosciences, Franklin Lakes, NJ, USA) at 37°C and 5% CO2. Cells were passaged once they reached 80–90% confluence. The medium was replenished every 2 days.
CLDN1 silencing
To determine the biological functions of CLDN1 in NPC, NPC-TW01 cells exhibiting silenced CLDN1 expression were constructed. NPC-TW01 cells were seeded into 24-well plates at a density of 1×105 cells/well. NPC-TW01 cells were divided into a small interfering RNA (siR)-negative control (NC) group and an siR-CLDN1 group (25 pmol/µl; Hanbio Biotechnology Co., Ltd., Shanghai, China). Cells in the siR-CLDN1 group were transfected with LV-GFP-PURO-siR-CLDN1 lentivirus (MOI=20; Hanbio Biotechnology Co., Ltd.) using Lipofectamine 3000 (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Following 12 h cultivation, RPMI-1640 medium was replenished. Subsequently, cells in siR-NC and siR-CLDN1 groups were transfected with siR-NC and siR-β-catenin (rescue group; 25 pmol/µl; Hanbio Biotechnology Co., Ltd.), respectively. On the next day, RPMI-1640 medium was replenished. Following 48 h of transfection, the cells were used for subsequent experiments.
CDN1 overexpression
NPC-TW01 cells were divided into an NC group and a CLDN1 group and transfected with a LV-GFP-PURO-CLDN1 lentivirus. Following 12 h cultivation, RPMI-1640 medium was replenished. Subsequently, cells in the NC and CLDN1 groups were transfected with pcDNA3.1-NC and pcDNA3.1-β-catenin (rescue group; 0.5 µg DNA; Hanbio Biotechnology Co., Ltd.), respectively. On the next day, RPMI-1640 medium was replenished. Following 48 h of transfection, the cells were used for subsequent experiments.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Cells were trypsinized and lysed with 1 ml TRIzol (Thermo Fisher Scientific Inc., Waltham, MA, USA). Following lysis, total RNA was extracted using the phenol chloroform method (27). RNA purity was determined at A260/A280 using an ultraviolet spectrophotometer (Nanodrop ND1000; Thermo Scientific, Inc.). cDNA was obtained from 1 µg RNA following reverse transcription with a PrimeScript RT Reagent kit (Takara Biotechnology, Co., Ltd., Dalian, China) and stored at −20°C. qPCR was performed using a SYBR Green RT-qPCR kit (Takara Biotechnology Co., Ltd.) and GAPDH was used as an internal reference. The qPCR system (20 µl) consisted of 10 µl SYBR EX Taq-Mix, 0.5 µl forward primer (CLDN1, 5′-CTGGGAGGTGCCCTACTTTG-3′; GAPDH, 5′-AGAAGGCTGGGGCTCATTTG-3′), 0.5 µl reverse primer (CLDN1, 5′-ACACGTAGTCTTTCCCGCTG-3′; GAPDH, 5′-AGGGGCCATCCACAGTCTTC-3′), 1 µl cDNA and 8 µl ddH2O. qPCR was performed in triplicate for each sample. The thermocycling conditions for qPCR were: Initial denaturation at 95°C for 5 min; 40 cycles of denaturation at 95°C for 1 min, annealing at 60°C for 30 sec, elongation at 72°C for 20 sec and final extension at 72°C for 1 min. The 2−ΔΔCq method was used to determine the relative expression of mRNA (28).
Western blotting
At 72 h after transfection, cells (1×106) were mixed with precooled radio immunoprecipitation assay lysis buffer (1 ml; 50 mM Tris-base, 1 mM EDTA, 150 mM NaCl, 0.1% sodium dodecyl sulfate, 1% Triton X-100 and 1% sodium deoxycholate; Beyotime Institute of Biotechnology, Shanghai, China) and lysed for 30 min on ice. Following sonication in an ice bath performed three times each at 5 sec (with a 20 sec interval), the mixture was kept on ice for 1 min and centrifuged at a speed of 12,000 × g at 4°C for 15 min. The supernatant was used to determine protein concentration using a bicinchoninic acid protein concentration determination kit [RTP7102; Real-Times (Beijing) Biotechnology Co., Ltd., Beijing, China]. Protein samples (20 µg/lane) were then mixed with 5X SDS loading buffer prior to denaturation in a boiling water bath for 10 min. Samples were then subjected to 10% SDS-PAGE at 100 V. Subsequently, resolved proteins were transferred to polyvinylidene difluoride membranes on ice (300 mA, 2 h). Following blocking with 50 g/l skim milk at room temperature for 1 h, membranes were incubated with polyclonal rabbit anti-human E-cadherin 1 (1:1,000; cat. no. ab15148; Abcam, Cambridge, UK), polyclonal rabbit anti-human vimentin (1:2,000; cat. no. ab45939; Abcam), polyclonal rabbit anti-human CLDN1 (1:1,000; cat. no. ab15098; Abcam), polyclonal rabbit anti-human β-catenin (1:1,000; cat. no. AF0066; Beyotime Institute of Biotechnology), monoclonal mouse anti-human H3 (1:1,000; cat. no. AF0009; Beyotime Institute of Biotechnology), and monoclonal mouse anti-human GAPDH (1:4,000; cat. no. AF0006; Beyotime Institute of Biotechnology) primary antibodies at 4°C overnight. Following 5 washes with phosphate-buffered saline with Tween 20 (5 min/wash), membranes were incubated with goat anti-rabbit (cat. no. A0208; for target proteins) or goat anti-mouse (cat. no. A0216; for GAPDH) horseradish peroxidase-conjugated secondary antibodies (1:3,000; Beyotime Institute of Biotechnology) for 1 h at room temperature. Membranes were then washed 5 times with phosphate-buffered saline with Tween-20 (5 min/wash). Membranes were developed using an enhanced chemiluminescence detection kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Image lab software v3.0 (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to acquire and analyze imaging signals. The expression of E-cadherin 1, vimentin CLDN1 and β-catenin were expressed relative to that of GAPDH, while nucleo-β-catenin was expressed relative to that of H3.
Cell-counting kit 8 (CCK-8) assay
All cells were seeded in 96-well plates at a density of 3,000/well. At 24, 48 and 72 h, the medium was discarded and cells were washed twice with phosphate-buffered saline, followed by the addition of RPMI-1640 medium (BD Biosciences) and 10% CCK-8 reaction reagent (Beyotime Institute of Biotechnology). Following incubation at 37°C for 1 h, the absorbance of each well was measured at 450 nm for plotting cell proliferation curves. Each sample was tested in triplicate.
Cell migration and invasion assays
Transwell chambers (8 µm diameter and 24 wells; Corning Inc., Corning, NY, USA) were used to evaluate the migration ability of cells. Cell suspension (200 µl; 2×105 cells) in serum-free RPMI-1640 medium (BD Biosciences) was added to the upper chamber. In the lower chamber, 500 µl RPMI-1640 medium supplemented with 10% fetal bovine serum was added. Following 24 h incubation, cells in the upper chamber were wiped using a cotton swab. Then, the chamber was fixed using 4% formaldehyde for 10 min at room temperature and then stained using Giemsa stain at room temperature for 1 min. Following 3 washes, cells that migrated to the other side of the chamber were counted in five fields using a light microscope (magnification, ×100) to evaluate migration ability.
The same number of cells as the migration assay were seeded in the upper chamber for the invasion assay. Matrigel (BD Biosciences) was used to determine the invasive ability of cells. Matrigel was diluted with serum-free RPMI-1640 medium at a ratio of 1:1. In the upper chamber, 100 µl diluted Matrigel was added and kept at 37°C for 1 h. In the lower chamber, 500 µl RPMI-1640 medium supplemented with 10% fetal bovine serum was added. Following 72 h incubation, cells in the upper chamber were wiped using cotton swab. The chamber was fixed using 4% formaldehyde for 10 min at room temperature and then stained using Giemsa stain at room temperature for 1 min. Following 3 washes, cells that moved to the other side of the chamber were counted using a microscope to evaluate invasive ability.
Statistical analysis
Results were analyzed using SPSS 17.0 statistical software (IBM Corp., Armonk, NY, USA). The data were expressed as the mean ± standard deviation. Differences between two groups were compared using Student's t-test and those among multiple groups were compared using one-way analysis of variance. Subsequently, a post hoc multiple comparison test (Tukey's honest significance test) was performed. P<0.05 was determined to indicate a statistically significant difference.
Results
CLDN1 expression is elevated in NPC cell lines
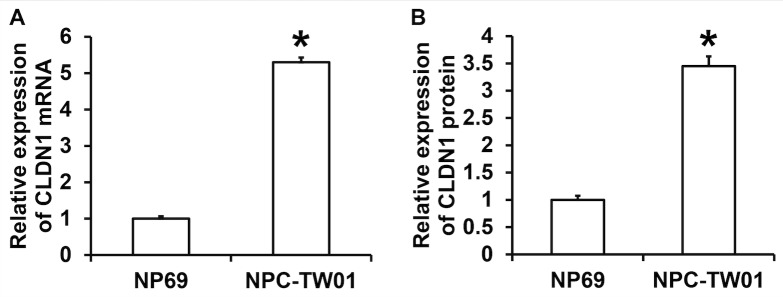
To determine the expression of CLDN1 in NPC cell lines, RT-qPCR and western blotting were performed. The results demonstrated that the expression of CLDN1 mRNA in NPC-TW01 cells was significantly higher than in NP69 cells (P<0.05; Fig. 1A). The results of western blotting demonstrated that the expression of CLDN1 protein in NPC-TW01 cells was significantly higher than in NP69 cells (P<0.05; Fig. 1B). These results demonstrate that CLDN1 expression is significantly elevated in NPC cells.
Figure 1.
Expression of CLDN1 (A) mRNA and (B) protein in NPC-TW01 cells compared with NP69 cells. Reverse transcription-quantitative polymerase chain reaction was used to determine CLDN1 mRNA levels, whereas protein expression was measured by western blotting. *P<0.05 vs. NP69 cells. CLDN1, Claudin-1.
CLDN1 silencing inhibits the proliferation of NPC cells whereas CLDN1 overexpression promotes the proliferation of NPC cells
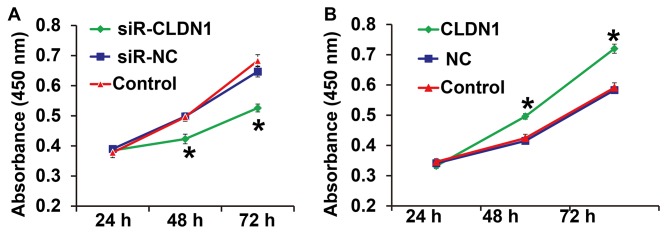
To measure cell proliferation, a CCK-8 assay was performed. The results demonstrated that the proliferation of NPC-TW01 cells transfected with siR-CLDN1 was significantly lower compared with cells in the siR-NC group at 48 and 72 h (P<0.05; Fig. 2A). By contrast, the proliferation of NPC-TW01 cells transfected with CLDN1 was significantly higher than that of cells in the NC group at 48 and 72 h (P<0.05; Fig. 2B). These results demonstrate that CLDN1 silencing inhibits the proliferation of NPC cells, whereas CLDN1 overexpression promotes the proliferation of NPC cells.
Figure 2.
NPC-TW01 cell proliferation following transfection with (A) siR-CLDN1 and (B) CLDN1. A Cell Counting Kit-8 assay was used to measure absorbance at 450 nm and proliferation curves were then plotted. *P<0.05 vs. siR-NC or NC group. CLDN1, claudin-1; siR, short hairpin RNA; NC, negative control.
CLDN1 promotes the migration and invasion of NPC cells
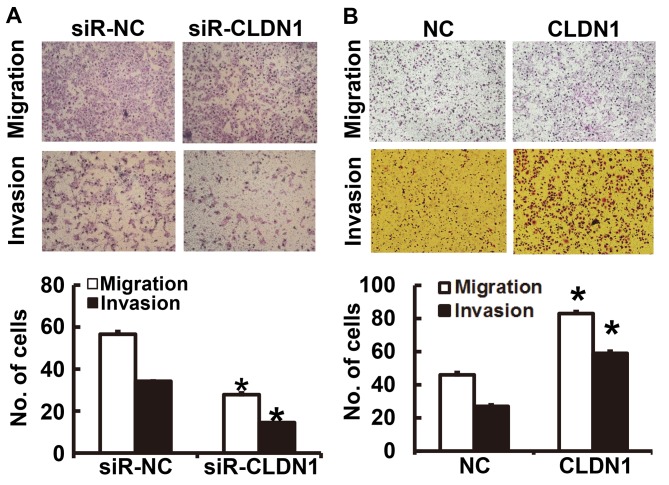
To determine the migration and invasion of NPC cells, a Transwell assay was employed. The results of the migration and invasion assays demonstrated that the number of NPC-TW01 cells transfected with siR-CLDN1 that passed through the Transwell chamber membrane was significantly lower than that of the siR-NC group (P<0.05; Fig. 3A). By contrast, the number of NPC-TW01 cells overexpressing CLDN1 that passed through the Transwell chamber membrane was significantly higher than those in the NC group (P<0.05; Fig. 3B). These results suggest that CLDN1 promotes the migration and invasion of NPC cells.
Figure 3.
The migration and invasion of NPC-TW01 cells transfected with (A) siR-CLDN1 and (B) CLDN1. Magnification, ×100. *P<0.05 vs. the respective siR-NC or NC groups. CLDN1, Claudin-1; siR, short hairpin RNA; NC, negative control.
CLDN1 overexpression promotes the EMT in NPC cells
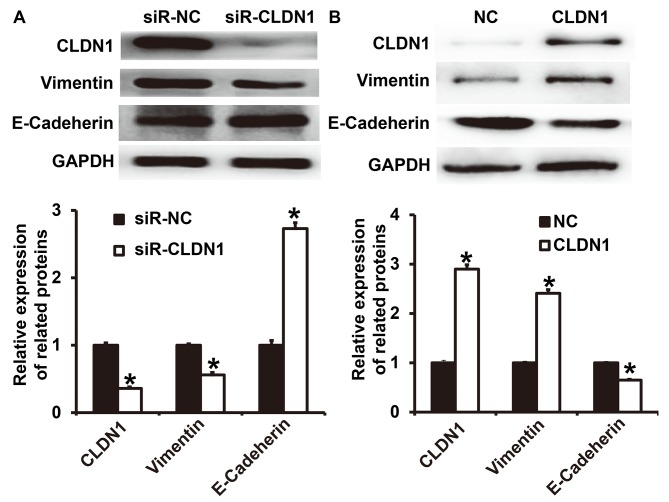
To determine the expression of the two EMT markers E-cadherin and vimentin following transfection of NPC-TW01 cells with either siR-CLDN1 or CLDN1, western blotting was performed. The results demonstrated that the expression of vimentin following transfection with siR-CLDN1 was significantly lower than that of the siR-NC group (P<0.05; Fig. 4A), however, the expression of E-cadherin following transfection with siR-CLDN1 was significantly higher than that of the siR-NC group (P<0.05). By contrast, the expression of vimentin following transfection with CLDN1 was significantly higher than that of the NC group (P<0.05; Fig. 4B), however, the expression of E-cadherin following transfection with CLDN1 was significantly lower than that of the NC group (P<0.05). These results indicate that the overexpression of CLDN1 promotes the EMT in NPC cells.
Figure 4.
Expression of vimentin and E-cadherin in NPC-TW01 cells transfected with (A) siR-CLDN1 and (B) CLDN1. *P<0.05 vs. siR-NC or NC group. CLDN1, claudin-1; siR, short hairpin RNA; NC, negative control.
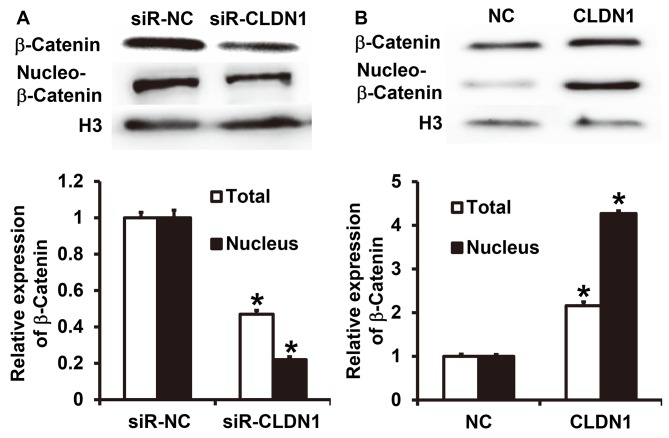
CLDN1 promotes β-catenin expression and entry into the nucleus and induces the EMT via the β-catenin signaling pathway
To measure the expression of β-catenin, western blotting was performed. The results demonstrated that CLDN1 silencing significantly inhibited the total expression of β-catenin and its nuclear translocation in NPC-TW01 cells (P<0.05; Fig. 5A). By contrast, overexpression of CLDN1 significantly enhanced the expression of β-catenin and its nuclear translocation in NPC-TW01 cells (P<0.05; Fig. 5B). These results suggest that CLDN1 promotes β-catenin expression and entry into the nucleus, and facilitates the EMT via the β-catenin signaling pathway.
Figure 5.
Expression of β-catenin in NPC-TW01 cells transfected with (A) siR-CLDN1 or (B) CLDN1. *P<0.05 vs. siR-NC or NC group. siR, short hairpin RNA; CLDN1, claudin-1; NC, negative control.
CLDN1 exerts its oncogene function via the β-catenin signaling pathway
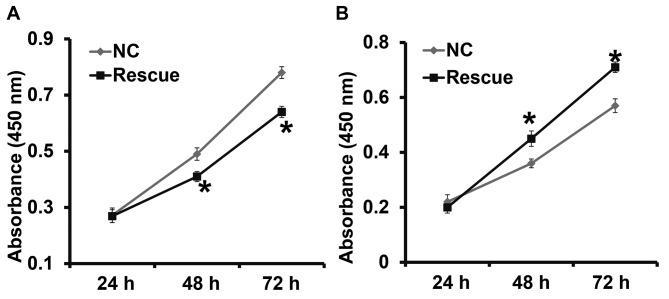
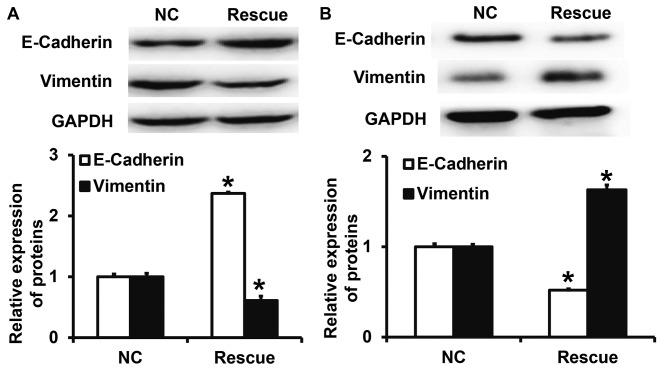
To confirm that CLDN1 induces its effects via the β-catenin signaling pathway, NPC-TW01 cells exhibiting CLDN1 silencing and overexpression were transfected with β-catenin siRNA and overexpression plasmids, respectively, to increase and decrease β-catenin expression (Fig. 6). The results demonstrated that the upregulation of β-catenin expression significantly promoted the proliferation (Fig. 7), invasion and migration (Fig. 8) and the EMT (Fig. 9) of NPC-TW01 cells with silenced CLDN1 expression. By contrast, downregulation of β-catenin expression inhibited the proliferation, invasion and migration, and EMT of NPC-TW01 cells exhibiting CLDN1 overexpression (Figs. 6–9). These results indicate that CLDN1 exerts its oncogene function via the β-catenin signaling pathway.
Figure 6.
Expression of β-catenin in the cytoplasm and nucleus of NPC-TW01 cells transfected with (A) siR-CLDN1 or (B) CLDN1. *P<0.05 vs. the respective NC. siR, short hairpin RNA; CLDN1, claudin-1; NC, negative control.
Figure 7.
Proliferation of NPC-TW01 cells transfected with β-catenin (A) siRNA or (B) overexpression plasmids. NPC-TW01 cells in siR-NC and siR-CLDN1 groups were transfected with siR-NC and siR-β-catenin (rescue group), respectively. NPC-TW01 cells in the NC and CLDN1 groups were transfected with pcDNA3.1-NC and pcDNA3.1-β-Catenin (rescue group), respectively. A Cell Counting Kit-8 assay was used to measure proliferation at absorbance at 450 nm and proliferation curves were then plotted. *P<0.05 vs. the respective NC. si, small interfering; NC, negative control.
Figure 8.
The migration and invasion of NPC-TW01 cells transfected with β-catenin (A) siRNA or (B) overexpression plasmids. NPC-TW01 cells in siR-NC and siR-CLDN1 groups were transfected with siR-NC and siR-β-catenin (rescue group), respectively. NPC-TW01 cells in the NC and CLDN1 groups were transfected with pcDNA3.1-NC and pcDNA3.1-β-catenin (rescue group), respectively. Magnification, 100×. *P<0.05 vs. the respective NC. si, small interfering; CLDN1, claudin-1; NC negative control.
Figure 9.
Expression of vimentin and E-cadherin in NPC-TW01 cells transfected with β-catenin (A) siRNA or (B) overexpression plasmids. NPC-TW01 cells in the siR-NC and siR-CLDN1 groups were transfected with siR-NC and siR-β-Catenin (rescue group), respectively. In addition, NPC-TW01 cells in NC and CLDN1 groups were transfected with pcDNA3.1-NC and pcDNA3.1-β-catenin (rescue group), respectively. *P<0.05 vs. the respective NC. si, small interfering; CLDN1, claudin-1; NC, negative control.
Discussion
CLDN1 is a tumor marker that exhibits abnormal expression in various different tumors and participates in the regulation of biological processes, including cell proliferation, apoptosis, invasion and migration (29). Nakagawa et al (30) indicated that CLDN1 overexpression promotes the invasion and migration of colon cancer cells and is negatively correlated with patient prognosis. Fortier et al (31) demonstrated that deletion of the keratin 8 and 18 genes upregulates the expression of CLDN1, thus stimulating the proliferation, invasion and migration of HepG2 cells. Jian et al (32) indicated that the function of CLDN1 in promoting the invasion and migration of osteosarcoma cells is closely associated with its detachment from the cell membrane and entry into the nucleus, suggesting that the intracellular location of CLDN1 is associated with tumor migration and invasion. It has also been reported that the expression of CLDN1 is elevated in gastric cancer tissues and that it inhibits the anoikis of gastric cancer cells via the β-catenin signaling pathway (33). These studies suggest that CLDN1 is closely associated with tumor invasion and metastasis and that the EMT is a key process in the migration of epithelial tumor cells. Certain studies have demonstrated that CLDN1 is closely associated with the EMT. For example, CLDN1 promotes the EMT in hepatocytes via the c- Abelson murine leukemia viral oncogene homolog 1-extracellular-signal-regulated kinase signaling pathway (34). In addition, the downregulation of CLDN1 facilitates the EMT of rat hepatocytes induced by transforming growth factor β (35). The function of CLDN1 in the EMT may differ among different cells. The results of the present study demonstrate that CLDN1 expression is upregulated in NPC cell lines and promotes the proliferation, the EMT, invasion and migration of NPC cells, which is consistent with its effects in other tumors.
As a type of multifunctional protein, β-catenin is widely distributed in different types of cells, including epithelial cells, fibroblasts and osteoblasts, and promotes the proliferation, differentiation and apoptosis of these cells (35). It has been demonstrated that the expression of β-catenin is upregulated in different types of tumor and promotes the EMT in these tumor cells, indicating that it is a key molecular target for inhibiting tumor metastasis. Oridonin inhibits the EMT in pancreatic cancer cells by downregulating the activity of the Wnt/β-catenin signaling pathway (36). Furthermore, the long non-coding RNA UCA1 promotes the EMT in breast cancer cells by activating the Wnt/β-catenin pathway (26) and Yi et al (37) determined that Wnt/β-catenin promotes the EMT and induces chemotherapy resistance in glioma. Wnt/β-catenin is not only a key signaling pathway that promotes the EMT, but also regulates tumor cell proliferation. Santos et al (38) reported that Sox9 enhances the proliferation of gastric cancer cells by activating the Wnt/β-catenin pathway. Furthermore, Lu et al (39) indicated that karyopherin β1 promotes the proliferation of glioma cells by activating the Wnt/β-catenin pathway. The results of these studies suggest that the Wnt/β-catenin signaling pathway induces important regulatory effects on the EMT and tumor proliferation. The results of the present study demonstrated that downregulating and overexpressing CLDN1 in NPC cells upregulates and downregulates the expression and nuclear entry of β-catenin, respectively. The downregulation of β-catenin inhibits the cancer-promoting function of CLDN1, suggesting that CLDN1 promotes the proliferation, EMT, invasion and migration of NPC cells by activating the Wnt/β-catenin signaling pathway.
In conclusion, the results of the present study demonstrate that CLDN1 promotes the proliferation, EMT, invasion and metastasis of NPC cells by activating the Wnt/β-catenin signaling pathway. Therefore, CLDN1 is an oncogene that may be a potential molecular therapeutic target for treating NPC.
Acknowledgements
The present study was supported by West China Hospital, Sichuan University, China. The authors wish to thank Professor Ping Li for his help.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
The final version of the manuscript has been read and approved by all authors, and each author believes that the manuscript represents honest work. XW, JX, CZ and YJ collaborated to design the study. XW, JX, CZ, CJZ, ZH, FW, YY and FF were responsible for the experiments. XW, JX, CZ and YJ analyzed the data. All authors collaborated to interpret results and develop the manuscript.
Ethics approval and consent to participate
All procedures performed in the current study were approved by the Ethics Committee of Sichuan University. Written informed consent was obtained from all patients or their families.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chen F, Chen C, Qu Y, Xiang H, Ai Q, Yang F, Tan X, Zhou Y, Jiang G, Zhang Z. Selenium-binding protein 1 in head and neck cancer is low-expression and associates with the prognosis of nasopharyngeal carcinoma. Medicine (Baltimore) 2016;95:e4592. doi: 10.1097/MD.0000000000004592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang HZ, Cao CN, Luo JW, Yi JL, Huang XD, Zhang SP, Wang K, Qu Y, Xiao JP, Li SY, et al. High-risk factors of parotid lymph node metastasis in nasopharyngeal carcinoma: A case-control study. Radiat Oncol. 2016;11:113. doi: 10.1186/s13014-016-0691-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang M, Wei S, Su L, Lv W, Hong J. Prognostic significance of pretreated serum lactate dehydrogenase level in nasopharyngeal carcinoma among Chinese population: A meta-analysis. Medicine (Baltimore) 2016;95:e4494. doi: 10.1097/MD.0000000000004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sung WW, Chu YC, Chen PR, Liao MH, Lee JW. Positive regulation of HIF-1A expression by EBV oncoprotein LMP1 in nasopharyngeal carcinoma cells. Cancer Lett. 2016;382:21–31. doi: 10.1016/j.canlet.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 5.Lin PJ, Twu CW, Liu YC, Lin TY, Wang WY, Lin JC. Comparison the clinical outcomes with altered versus conventional fractionated radiotherapy plus concurrent chemotherapy for advanced nasopharyngeal carcinoma. Head Neck. 2018 Feb 1; doi: 10.1002/hed.25084. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 6.Zhang L, Huang Y, Hong S, Yang Y, Yu G, Jia J, Peng P, Wu X, Lin Q, Xi X, et al. Gemcitabine plus cisplatin versus fluorouracil plus cisplatin in recurrent or metastatic nasopharyngeal carcinoma: A multicentre, randomised, open-label, phase 3 trial. Lancet. 2016;388:1883–1892. doi: 10.1016/S0140-6736(16)31388-5. [DOI] [PubMed] [Google Scholar]
- 7.Shi P, Zhong J, Hong J, Huang R, Wang K, Chen Y. Automated Ki-67 quantification of immunohistochemical staining image of human nasopharyngeal carcinoma xenografts. Sci Rep. 2016;6:32127. doi: 10.1038/srep32127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu VW, Leung WS, Wong KL, Chan YK, Law WL, Leung WK, Yu YL. The impact of positron emission tomography on primary tumour delineation and dosimetric outcome in intensity modulated radiotherapy of early T-stage nasopharyngeal carcinoma. Radiat Oncol. 2016;11:109. doi: 10.1186/s13014-016-0685-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou ZW, Ma C, Medoro L, Chen L, Wang B, Gupta R, Liu T, Yang XZ, Chen TT, Wang RZ, et al. LncRNA ANRIL is up-regulated in nasopharyngeal carcinoma and promotes the cancer progression via increasing proliferation, reprograming cell glucose metabolism and inducing side-population stem-like cancer cells. Oncotarget. 2016;7:61741–61754. doi: 10.18632/oncotarget.11437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearce SC, Al-Jawadi A, Kishida K, Yu S, Hu M, Fritzky LF, Edelblum KL, Gao N, Ferraris RP. Marked differences in tight junction composition and macromolecular permeability among different intestinal cell types. BMC Biol. 2018;16:19. doi: 10.1186/s12915-018-0481-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez-Mariscal L, Posadas-Torrentera Y, Miranda J, Uc P, Ortega-Olvera JM, Hernandez S. Strategies that target tight junctions for enhanced drug delivery. Curr Pharm Des. 2016;22:1305–1311. doi: 10.2174/1381612822666160720163656. [DOI] [PubMed] [Google Scholar]
- 12.Salvador E, Burek M, Forster CY. Tight junctions and the tumor microenvironment. Curr Pathobiol Rep. 2016;4:135–145. doi: 10.1007/s40139-016-0106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaouche-Mazouni S, Scherpereel A, Zaamoum R, Mihalache A, Amir ZC, Lebaïli N, Delaire B, Gosset P. Claudin 3, 4 and 15 expression in solid tumors of lung adenocarcinoma versus malignant pleural mesothelioma. Ann Diagn Pathol. 2015;19:193–197. doi: 10.1016/j.anndiagpath.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Kudinov AE, Deneka A, Nikonova AS, Beck TN, Ahn YH, Liu X, Martinez CF, Schultz FA, Reynolds S, Yang DH, et al. Musashi-2 (MSI2) supports TGF-β signaling and inhibits claudins to promote non-small cell lung cancer (NSCLC) metastasis; Proc Natl Acad Sci USA; 2016; pp. 6955–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon MJ, Kim SH, Jeong HM, Jung HS, Kim SS, Lee JE, Gye MC, Erkin OC, Koh SS, Choi YL, et al. Claudin-4 overexpression is associated with epigenetic derepression in gastric carcinoma. Lab Invest. 2011;91:1652–1667. doi: 10.1038/labinvest.2011.117. [DOI] [PubMed] [Google Scholar]
- 16.Okugawa T, Oshima T, Chen X, Hori K, Tomita T, Fukui H, Watari J, Matsumoto T, Miwa H. Down-regulation of claudin-3 is associated with proliferative potential in early gastric cancers. Dig Dis Sci. 2012;57:1562–1567. doi: 10.1007/s10620-012-2043-5. [DOI] [PubMed] [Google Scholar]
- 17.Troy TC, Arabzadeh A, Yerlikaya S, Turksen K. Claudin immunolocalization in neonatal mouse epithelial tissues. Cell Tissue Res. 2007;330:381–388. doi: 10.1007/s00441-007-0487-2. [DOI] [PubMed] [Google Scholar]
- 18.Katoh M, Katoh M. CLDN23 gene, frequently down-regulated in intestinal-type gastric cancer, is a novel member of CLAUDIN gene family. Int J Mol Med. 2003;11:683–689. [PubMed] [Google Scholar]
- 19.Chen JJ, Zhong M, Dou TH, Wu ZY, Tang WJ. rs17501976 polymorphism of CLDN1 gene is associated with decreased risk of colorectal cancer in a Chinese population. Int J Clin Exp Med. 2015;8:1247–1252. [PMC free article] [PubMed] [Google Scholar]
- 20.Benczik M, Galamb A, Koiss R, Kovács A, Járay B, Székely T, Szekerczés T, Schaff Z, Sobel G, Jeney C. Claudin-1 as a biomarker of cervical cytology and histology. Pathol Oncol Res. 2016;22:179–188. doi: 10.1007/s12253-015-9990-z. [DOI] [PubMed] [Google Scholar]
- 21.Hsueh C, Chang YS, Tseng NM, Liao CT, Hsueh S, Chang JH, Wu IC, Chang KP. Expression pattern and prognostic significance of claudins 1, 4 and 7 in nasopharyngeal carcinoma. Hum Pathol. 2010;41:944–950. doi: 10.1016/j.humpath.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 22.Cantelli G, Crosas-Molist E, Georgouli M, Sanz-Moreno V. TGFB-induced transcription in cancer. Semin Cancer Biol. 2017;42:42–62. doi: 10.1016/j.semcancer.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cardenas H, Zhao J, Vieth E, Nephew KP, Matei D. EZH2 inhibition promotes epithelial-to-mesenchymal transition in ovarian cancer cells. Oncotarget. 2016;7:84453–84467. doi: 10.18632/oncotarget.11497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin L, Yi J, Gao Y, Han S, He Z, Chen L, Song H. MiR-630 inhibits invasion and metastasis in esophageal squamous cell carcinoma. Acta Biochim Biophys Sin (Shanghai) 2016;48:810–819. doi: 10.1093/abbs/gmw073. [DOI] [PubMed] [Google Scholar]
- 25.Chung YH, Li SC, Kao YH, Luo HL, Cheng YT, Lin PR, Tai MH, Chiang PH. MiR-30a-5p inhibits epithelial-to-mesenchymal transition and upregulates expression of tight junction protein claudin-5 in human upper tract urothelial carcinoma cells. Int J Mol Sci. 2017;18:E1826. doi: 10.3390/ijms18081826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao C, Wu CH, Hu HZ. LncRNA UCA1 promotes epithelial-mesenchymal transition (EMT) of breast cancer cells via enhancing Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2016;20:2819–2824. [PubMed] [Google Scholar]
- 27.Cepollaro S, Della Bella E, de Biase D, Visani M, Fini M. Evaluation of RNA from human trabecular bone and identification of stable reference genes. J Cell Physiol. 2017;233:4401–4407. doi: 10.1002/jcp.26319. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.De Vicente JC, Fernandez-Valle A, Vivanco-Allende B, Santamarta TR, Lequerica-Fernández P, Hernández-Vallejo G, Allonca-Campa E. The prognostic role of claudins −1 and −4 in oral squamous cell carcinoma. Anticancer Res. 2015;35:2949–2959. [PubMed] [Google Scholar]
- 30.Nakagawa S, Miyoshi N, Ishii H, Mimori K, Tanaka F, Sekimoto M, Doki Y, Mori M. Expression of CLDN1 in colorectal cancer: A novel marker for prognosis. Int J Oncol. 2011;39:791–796. doi: 10.3892/ijo.2011.1102. [DOI] [PubMed] [Google Scholar]
- 31.Fortier AM, Asselin E, Cadrin M. Keratin 8 and 18 loss in epithelial cancer cells increases collective cell migration and cisplatin sensitivity through claudin1 up-regulation. J Biol Chem. 2013;288:11555–11571. doi: 10.1074/jbc.M112.428920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jian Y, Chen C, Li B, Tian X. Delocalized Claudin-1 promotes metastasis of human osteosarcoma cells. Biochem Biophys Res Commun. 2015;466:356–361. doi: 10.1016/j.bbrc.2015.09.028. [DOI] [PubMed] [Google Scholar]
- 33.Huang J, Zhang L, He C, Qu Y, Li J, Zhang J, Du T, Chen X, Yu Y, Liu B, Zhu Z. Claudin-1 enhances tumor proliferation and metastasis by regulating cell anoikis in gastric cancer. Oncotarget. 2015;6:1652–1665. doi: 10.18632/oncotarget.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suh Y, Yoon CH, Kim RK, Lim EJ, Oh YS, Hwang SG, An S, Yoon G, Gye MC, Yi JM, et al. Claudin-1 induces epithelial-mesenchymal transition through activation of the c-Abl-ERK signaling pathway in human liver cells. Oncogene. 2013;32:4873–4882. doi: 10.1038/onc.2012.505. [DOI] [PubMed] [Google Scholar]
- 35.Kojima T, Takano K, Yamamoto T, Murata M, Son S, Imamura M, Yamaguchi H, Osanai M, Chiba H, Himi T, Sawada N. Transforming growth factor-beta induces epithelial to mesenchymal transition by down-regulation of claudin-1 expression and the fence function in adult rat hepatocytes. Liver Int. 2008;28:534–545. doi: 10.1111/j.1478-3231.2007.01631.x. [DOI] [PubMed] [Google Scholar]
- 36.Liu QQ, Chen K, Ye Q, Jiang XH, Sun YW. Oridonin inhibits pancreatic cancer cell migration and epithelial-mesenchymal transition by suppressing Wnt/β-catenin signaling pathway. Cancer Cell Int. 2016;16:57. doi: 10.1186/s12935-016-0336-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yi GZ, Liu YW, Xiang W, Wang H, Chen ZY, Xie SD, Qi ST. Akt and β-catenin contribute to TMZ resistance and EMT of MGMT negative malignant glioma cell line. J Neurol Sci. 2016;367:101–106. doi: 10.1016/j.jns.2016.05.054. [DOI] [PubMed] [Google Scholar]
- 38.Santos JC, Carrasco-Garcia E, Garcia-Puga M, Aldaz P, Montes M, Fernandez-Reyes M, de Oliveira CC, Lawrie CH, Araúzo-Bravo MJ, Ribeiro ML, Matheu A. SOX9 elevation acts with canonical WNT signaling to drive gastric cancer progression. Cancer Res. 2016;76:6735–6746. doi: 10.1158/0008-5472.CAN-16-1120. [DOI] [PubMed] [Google Scholar]
- 39.Lu T, Bao Z, Wang Y, Yang L, Lu B, Yan K, Wang S, Wei H, Zhang Z, Cui G. Karyopherinβ1 regulates proliferation of human glioma cells via Wnt/β-catenin pathway. Biochem Biophys Res Commun. 2016;478:1189–1197. doi: 10.1016/j.bbrc.2016.08.093. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.