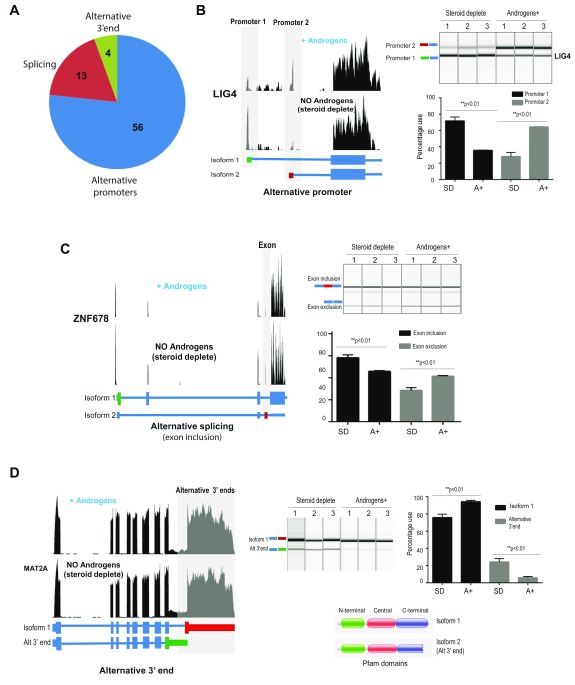
Figure 1. Global identification of androgen-dependent mRNA isoform production in prostate cancer cells predicts a major role for alternative promoter utilisation.
(A) Analysis of RNAseq data from LNCaP cells grown with (A+) or without androgens (R1881) (steroid deplete, SD) for 24 hours identified 73 androgen regulated alternative mRNA isoforms. The 73 alternative events were generated via androgen-regulated utilisation of 56 alternative promoters, 4 alternative 3' ends and 13 alternative splicing events. (B) Androgens drive a promoter switch in the LIG4 gene, which produces an mRNA isoform with an alternative 5’UTR. Visualisation of our LNCaP cell RNA-seq reads for the LIG4 gene on the UCSC genome browser identified a switch from promoter 1 to alternative promoter 2 in cells grown in the presence of androgens. Promoter 2 is predicted to produce a different 5’UTR without influencing the protein sequence (left panel). Quantitative PCR using primers specific to each promoter indicate that in response to androgens there is repression of promoter 1 and induction of promoter 2 (right panel). (C) Androgens drive alternative splicing of the ZNF678 gene. Visualisation of our LNCaP cell RNA-seq reads for the ZNF678 gene on the UCSC genome browser identified a switch to inclusion of a cassette exon in the presence of androgens. Inclusion of the alternative cassette exon in the ZNF678 gene is predicted to induce a switch to an alternative non-coding mRNA isoform (left panel). Quantitative PCR using primers in flanking exons confirmed increased inclusion of the alternative exon in LNCaP cells exposed to androgens (right panel). (D) Androgens promote selection of an alternative 3’ end for the MAT2A gene. Visualisation of our LNCaP cell RNA-seq reads for the MAT2A gene on the UCSC genome browser indicates a switch to reduced usage of an alternative 3’ end in the presence of androgens (left panel). Quantitative PCR using primers specific to each isoform confirmed down-regulation of an alternative 3’ end (p<0.01). Alternative 3’ ends for the MAT2A gene are predicted to produce proteins with different amino acid sequences and to influence a known Pfam domain (right panel).