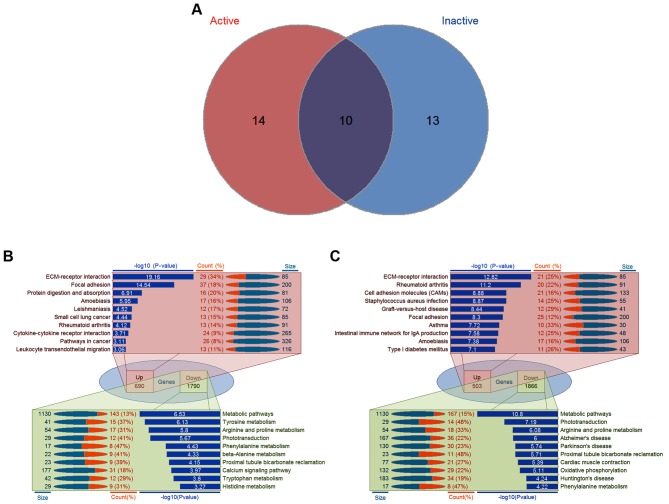
Figure 2.
Significantly enriched pathways associated with PDR. (A) Venn diagram illustrating the overlapping pathways in the active and inactive samples. (B) Detailed information of the top 10 pathways in the active samples. For 690 upregulated genes in the active FVMs, the top 10 pathways were ECM-receptor interaction, focal adhesion, protein digestion and absorption, amoebiasis, leishmaniasis, small cell lung cancer, rheumatoid arthritis, cytokine-cytokine receptor interaction, pathways in cancer, and leukocyte transendothelial migration; for 1,790 downregulated genes in the active FVMs, the top 10 pathways were metabolic pathways, tyrosine metabolism, arginine and proline metabolism, phototransduction, phenylalanine metabolism, β-alanine metabolism, proximal tubule bicarbonate reclamation, calcium signaling pathway, tryptophan metabolism, and histidine metabolism. (C) Detailed information of the top 10 pathways in the inactive samples. For 503 upregulated genes in the active FVMs, the top 10 pathways were ECM-receptor interaction, focal adhesion, protein digestion and absorption, amoebiasis, leishmaniasis, small cell lung cancer, rheumatoid arthritis, cytokine-cytokine receptor interaction, pathways in cancer, and leukocyte transendothelial migration; for 1,866 downregulated genes in the active FVMs, the top 10 pathways were metabolic pathways, tyrosine metabolism, arginine and proline metabolism, phototransduction, phenylalanine metabolism, β-alanine metabolism, proximal tubule bicarbonate reclamation, calcium signaling pathway, tryptophan metabolism, and histidine metabolism. PDR, proliferative diabetic retinopathy; FVMs, fibrovascular membranes; Up, upregulated; Down, downregulated; ECM, extracellular matrix.