Abstract
D Rhamnose β-hederin (DRβ-H), a novel oleanane-type triterpenoid saponin isolated from the traditional Chinese medicinal plant Clematis ganpiniana, has been demonstrated to be effective against various types of tumor. However, the exact role of DRβ-H on breast cancer remains largely unresolved. In the present study, it was observed that DRβ-H exhibited anti-proliferative and pro-apoptotic activity in human breast cancer cells (MCF-7/S). DRβ-H was able to inhibit exosome secretion, and the level of exosomes was positively associated with cell growth after absorption and internalization by target breast cancer cells. By analyzing the miRNA profiles of exosomes and MCF-7/S, it was identified that several miRNAs were detected exclusively in exosomes. Knockdown of the top five exosomal miRNAs and an MCF-7/S proliferation assay indicated that exosomal miR-130a and miR-425 may enhance MCF-7/S cell viability. Target gene prediction and pathway analysis revealed the involvement of miR-130a and miR-425 in pathways associated with malignant cell proliferation. These results demonstrated that DRβ-H inhibited MCF-7/S cell growth through reducing exosome release.
Keywords: breast cancer, exosomes, D Rhamnose β-Hederin, microRNAs
Introduction
Breast cancer is the most common malignant tumor type in women world wide (1). Although several therapeutic advances have been achieved in extending the disease-free survival and overall survival times of patients, treatments for breast cancer remain far from satisfactory. Identifying active components from natural plants has become a novel treatment strategy (2,3). Currently, numerous anti-cancer agents have been identified, including Clematis ganpiniana (4).
Clematis ganpiniana, in the Ranunculaceae family and Clematis genus, are widely used in traditional Chinese medicine. Although the roots and rhizomes of Clematis ganpiniana may be used as antibacterial and anticancer agents, the chemical components require additional study (4). In a previous study, four types of triterpenoid derivatives were identified and purified from Clematis ganpiniana, which were demonstrated to induce cytotoxicity in breast cancer cells (4). Furthermore, D Rhamnose β-hederin (3β-[(α-L-arabinopyranosyl)-oxy] olean-12-en-28-oicacid) (DRβ-H), one of the isolated oleanane-type triterpenoid saponins, was investigated. The apoptotic activity and anticancer properties of DRβ-H have been previously demonstrated, suggesting that DRβ-H may be a potential treatment for breast cancer (5). However, the underlying molecular mechanism by which DRβ-H inhibits malignant cells growth remains unclear.
Exosomes have attracted interest since the realization that these small vesicles are not merely cellular debris, but rather important regulators in tumor biology processes, including angiogenesis, invasiveness, metastasis and chemoresistance (6). They contain a wide variety of proteins, lipids and mRNAs, including microRNAs (miRNAs) that maybe transferred from donor cells to recipient cells inducing epigenetic changes (7,8). Exosome secretion is one of the mechanisms by which breast cancer cells communicate with surrounding cells and reprogram the tumor microenvironment (9,10). By using established cell lines (11), previous studies demonstrated that drug-resistant breast cancer cells may spread chemoresistance to target cells by releasing abundant exosomes and that such modulatory effects may be partly attributed to the intercellular transfer of specific miRNAs (12,13). In the present study, the effects of DRβ-H on exosome secretion and the functions of breast cancer-derived exosomes in cell growth were evaluated.
Materials and methods
Cell culture and drug preparation
The cell line used in the present study was wild-type drug-sensitive MCF-7 breast cancer cells (MCF-7/S) purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). In selected experiments, MCF-7/S expressing green fluorescent protein (GFP-S) was generated and characterized as previously reported (14). Cells were cultured in a 5% CO2 atmosphere at 37°C in Dulbecco's modified Eagle's medium with a high glucose content (HyClone; GE Healthcare Life Sciences, Logan, UT, USA), supplemented with 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100 U/ml penicillin and 100 µg/ml streptomycin. For routine passage, cultures were split 1:3 when they reached between 70 and 80% confluence every 2–3 days. To minimize the influence of exosomes in FBS, FBS was ultracentrifuged at 100,000 × g at 4°C overnight to spin down any potential vesicular content (Avanti J-30I; Beckman Coulter, Inc., Brea, CA, USA) and used for all studies. The protocols followed for the extraction, purification and analysis of DRβ-H are described in a previous study (4).
MTT assay
The inhibitory effect of DRβ-H on breast cancer cells was determined usingan MTT assay as previously described with minor modifications (5). Briefly, 5×103 cells were cultured in 96-well plates in triplicate. After 8 h, DRβ-H was added at 0, 5, 10, 20, 40 or 80 µg/ml. As a negative control, cells were treated with PBS. The cells were cultured for 0, 24, 48 and 72 h and then 20 µl MTT was added into each well at 5 mg/ml final concentration for 4 h. Following incubation, 150 µl dimethyl sulfoxide was added to dissolve the formazan crystals for 20 min at room temperature. The optical density value for each well was detected at a wavelength of 490 nm with a microplate reader (5082 Grodig; Tecan Group, Ltd., Mannedorf, Switzerland).
Flow cytometry analysis
Breast cancer cells exposed to the indicated concentrations of DRβ-H (0, 10, 20 or 40 µg/ml) for 24 h were harvested. Cells were washed twice with cold PBS, incubated with Annexin-V-FITC (BDBiosciences, Franklin Lakes, NJ, USA) and propidium iodide for 15 min at room temperature in the dark, and analyzed using a flow cytometer (BD FACS Calibur; BD Biosciences, Franklin Lakes, NJ, USA).
Exosome isolation and characterization
Exosome isolation and characterization was performed using protocols as previously described (12). Exosomes were lysed for protein/RNA extraction, or diluted in PBS for incubation assay or labeled for confocal observation (12). The morphology and size of exosomes were examined by transmission electron microscopy (JEM-1010; JEOL, Ltd., Tokyo, Japan) as previously reported (15).
Exosome uptake
Exosomes were stained with a PKH26 Red Fluorescent Cell Linker kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) according to a previously published protocol (12). As the negative control, exosomes without PKH26 labeling were also prepared. For observation of exosome uptake, confocal laser scanning microscopy (LSM710; Zeiss GmbH, Jena, Germany) was used (150,000 magnification).
RNA isolation and microarray
Exosomal RNA was isolated using the Total Exosome RNA and Protein Isolation kit (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in accordance with the manufacturer's protocol. Cellular RNA was prepared using TRIzol®reagent (Thermo Fisher Scientific, Inc.). miRNA profiles were analyzed using an Affymetrix Gene Chip miRNA Array 3.0 (Capital Bio Corporation, Beijing, China) according to the manufacturer's protocol. The levels of miRNAs between exosomes and MCF-7/S were calculated as previously reported (16).
Transfection of miRNA inhibitors
miRNA inhibitors and the negative controls were synthesized by GenePharma Co., Ltd. (Shanghai, China). Transfection of miRNA inhibitors was performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Cells and exosomes were harvested 48 h after transfection for subsequent analysis viapolymerase chain reaction (PCR).
Western blot analysis
Proteins were extracted from exosomes using the Total Exosome RNA and Protein Isolation kit (Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's protocols. Equal amounts of proteins were subjected to SDS-PAGE and transferred to polyvinylidenedi fluoride membranes. The CD63 protein level was measured by western blotting analysis using anantibody against CD63 (Santa Cruz, USA). β-actin (Sigma, Germany) was used as an internal control for normalization. Then, bound proteins were visualized using the ECL Plus kit (EMD Millipore, Billerica, MA, USA) with Image Lab Software version 5.2.1 (Bio-Rad, USA).
Reverse transcription-quantitative (RT-q) PCR
The RT-qPCR was run in triplicate using the SYBR green (Biouniquer Technology, Nanjing, China) technique. Briefly, cDNA for miRNA was synthesized using the BU-Script RT kit (Biouniquer Technology, Nanjing, China). Specific stem-loop primers (Springen Biotechnology, Nanjing, China) were designed for the selected miRNAs, and U6 was used as an internal control. Expressions of miRNAs and U6 were evaluated using following primers: miR-130a forward, 5′-TTCACATTGTGCTACTGTCTGC-3′; miR-183 forward, 5′-TATGGCACTGGTAGAATTCACT-3′; miR-20b forward, 5′-TGTCAACGATACGCTACGA-3′; miR-25 forward, 5′-TCTGGTCTCCCTCACAGGAC-3′; and miR-452 forward, 5′-GCGAACTGTTTGCAGAGG-3′. Gene amplifications were performed on a Light Cycler 480 (Roche, Australia) as follows: 91°C for 5 min followed by 45 cycles (91°Cf or 15 sec, and 60°C for 30 sec), followed by melting curve detection. The relative miRNA expression was calculated using the 2−ΔΔCq method.
Target gene prediction
The TargetScan (http://www.targetscan.org/) and miRDB (http://www.mirdb.org/miRDB/) databases were employed to predict miRNA targets (17,18). Only the genes predicted by these two independent tools were considered. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were analyzed by the online DAVID program (http://david.abcc.ncifcrf.gov/) (19,20).
Statistical analysis
Statistical analysis was performed using SPSS 20.0 statistical software (IBM Corp., Armonk, NY, USA). All experiments were performed in triplicate and the representative data are presented as the mean ± standard deviation. Differences were determined using the Student's t-test or using a one-way ANOVA with Student-Newman-Keuls post-hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
DRβ-H inhibits the proliferation of MCF-7/S cells
The chemical structure of DRβ-H is presented in Fig. 1A. MCF-7/S cells were treated with different concentrations of DRβ-H (0, 5, 10, 20, 40 or 80 µg/ml) or for different times (0, 24, 48 or 72 h) and the rate of inhibition of growth was measured using an MTT assay. DRβ-H exerted a significant inhibitory effect on MCF-7/S cells in a dose- and time-dependent manner compared with the negative control (Fig. 1B and C).
Figure 1.
Effects of DRβ-H on the proliferation and apoptosis of MCF-7/S cells. (A) The chemical structure of DRβ-H. (B) MTT assay of MCF-7/S treated by different concentrations of DRβ-H (0, 5, 10, 20, 40 and 80 µg/ml) for 24 h. DRβ-H inhibited cell proliferation in a dose-dependent manner. (C) MTT assay of MCF-7/S treated by 20 µg/ml DRβ-H for different times (0, 24, 48 and 72 h). DRβ-H inhibited cell proliferation in a time-dependent manner. (D) Flow cytometry analysis of MCF-7/S treated by different concentrations of DRβ-H (0, 10, 20 and 40 µg/ml) for 24 h. DRβ-H induced cell apoptosis in a dose-dependent manner. Data are presented as the mean ± the standard deviation of three independent experiments. *P<0.05 vs. DRβ-H-untreated group. The ** and # shows that the two groups had statistically significant difference (P<0.05). MCF-7/s, wild-type drug-sensitive MCF-7 breast cancer cells; DRβ-H, DRhamnose β-hederin.
DRβ-H induces apoptosis in MCF-7/S cells
To explore whether the anti-tumor effect of DRβ-H was also associated with apoptosis, MCF-7/S cells were treated with different concentrations of DRβ-H (0, 10, 20 or 40 µg/ml) for 24 h and the rate of apoptosis was determined by flow cytometry. Incubation of MCF-7/S cells with DRβ-H increased the apoptotic rate compared with control cells treated without DRβ-H (Fig. 1D). Furthermore, it was identified that increased DRβ-H concentrations increased the apoptotic effect, demonstrating a dose-dependent association.
DRβ-H reduces exosome release in MCF-7/S cells
The protocol of exosome isolation was based on differential sedimentation properties, and used a series of centrifugation and ultracentrifugation steps as previously published (12). Exosomes were homogeneous in morphology, and exhibited round vesicles measuring between 50 and 100 nm in diameter as determined by transmission electron microscopy (Fig. 2A). The exosome quantity was evaluated by measuring protein content. The authors' previous study demonstrated that exosomes derived from cancer cells were responsible for drug efficacy during toxic insult (13). Thus, it was hypothesized exosome release is involved in DRβ-H-mediated growth inhibition. MCF-7/S cells were treated with different concentrations of DRβ-H and it was revealed that the quantity of exosomes was decreased in a dose-dependent manner (Fig. 2B). Additionally, treatment of cells with high concentration of DRβ-H reduced exosome marker CD63 compared with the control cells treated with lower concentration of DRβ-H (Fig. 2C), indicating that DRβ-H may inhibit exosome secretion.
Figure 2.
DRβ-H reducesexosomerelease in MCF-7/S cells. (A) Transmission electron microscopic image of exosomes isolated from MCF-7/S cells. Bar indicates 200 nm. (B) Exosome quantity of MCF-7/S treated by different concentrations of DRβ-H (0, 5, 10, 20, 40 and 80 µg/ml). DRβ-H decreased exosome secretion in a dose-dependent manner. (C) Western blotting for CD63 expression in exosomes after MCF-7/S were treated by different concentrations of DRβ-H (0, 20 and 80 µg/ml). Data are presented as the mean ± the standard deviation of three independent experiments. MCF-7/s, wild-type drug-sensitive MCF-7 breast cancer cells; DRβ-H, DRhamnose β-hederin.
DRβ-H suppresses MCF-7/S growth by inhibiting exosome release
To visualize exosome uptake by recipient cells, exosomes were labeled by PKH26 red dye and incubated with GFP-S for 24 h, after which confocal laser scanning microscopy was performed. The internalization of exosomes was indicated by several red fluorescent punctuated signals on the cell membranes and inside the cytoplasm of GFP-S (Fig. 3A; white arrows indicate red fluorescent punctuated signals). The effects of exosomes on cell growth were assessed in DRβ-H-treated MCF-7/S cells. As presented in Fig. 3, the proliferation rate modulated by DRβ-H was relatively lower in MCF-7/S cells without exosomes, whereas a significant increase in proliferation rate was identified in MCF-7/S cells treated with exosomes (Fig. 3B). Furthermore, incubation of MCF-7/S cells with exosomes significantly decreased apoptosis when compared with control cells incubated without exosomes (Fig. 3C). These data, along with the observations in the aforementioned section, collectively suggested that exosomes were able to promote recipient MCF-7/S cells growth and DRβ-H suppressed MCF-7/S cells growth by inhibiting exosome release.
Figure 3.
DRβ-H suppresses MCF-7/S growth by inhibiting exosome release. (A) Confocal laser microscopic images of GFP-S exposed to PKH26-stainedexosomes for 24 h; (a) green signal from established GFP-S cells; (b) red fluorescent signal from PKH26-labeled exosomes; (c) merged image of (a) and (b) presents the internalization of exosomes (white arrows) on the cell membranes and inside the cytoplasm of GFP-S. (B) Different concentrations of exosomes from 0–200 µg/ml were added into DRβ-H-treated MCF-7/S cells. Exosomes promoted cell proliferation in a dose-dependent manner. (C) Different concentrations of exosomesfrom 0–200 µg/ml were added into DRβ-H-treated MCF-7/S cells. Exosomes decreased cell apoptosis in a dose-dependent manner. Data are presented as the mean ± the standard deviation of three independent experiments. *P<0.05 vs. exosome-untreated group. MCF-7/s, wild-type drug-sensitive MCF-7 breast cancer cells; DRβ-H, DRhamnose β-hederin.
DRβ-H decreases exosomal miRNAs to regulate MCF-7/S proliferation
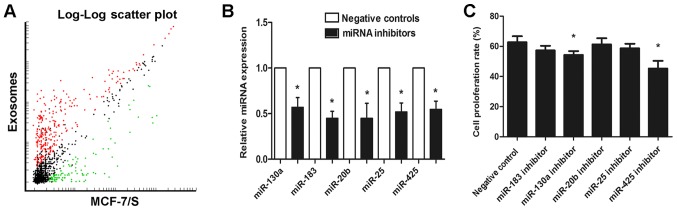
Given that exosomes contain cellular information, an attempt was made to explore whether exosomal miRNAs affect MCF-7/S proliferation. By comparing the miRNA profiles of exosomes and their cells of origin, it was identified that there are numerous miRNAs shared by exosomes and MCF-7/S. Furthermore, a number of miRNAs were detected exclusively in exosomes compared with those in the parental cells, whereas a group of miRNAs expressed by cells were absent in the corresponding exosomes (Fig. 4A). Next, the functions of the top five miRNAs (miR-130a, miR-183, miR-20b, miR-25 and miR-425) more localized in exosomes compared with that incells were examined by transfecting with miRNA inhibitors. The successful knockdown of the five exosomal miRNAs was subsequently validated by RT-qPCR (Fig. 4B). miRNA knockdown exosomes were added into recipient MCF-7/S cells and cell viability was detected by analyzing the proliferation rate. As presented in Fig. 4, of the five miRNAs tested, only miR-130a and miR-425 knockdown exosomes were able to significantly decrease cell proliferation (Fig. 4C), indicating that exosomal miR-130a and miR-425 could enhance MCF-7/s cell viability.
Figure 4.
DRβ-H decreases exosomal miRNAs to regulate MCF-7/S proliferation. (A) A scatter plot of miRNA signal intensity revealed that miRNA repertoires of exosomes were different from those of their parental cells. (B) Relative expressions of the top five exosomal miRNAs (miR-130a, miR-183, miR-20b, miR-25, and miR-425) following transfecting with the corresponding miRNA inhibitors. Negative control groups were transfected with the specific inhibitors, which have the same amount as the basic group but a different sequence from those miRNAs. (C) Effects of miRNA knockdown exosomes on MCF-7/S proliferation. Data are presented as the mean ± the standard deviation of three independent experiments. *P<0.05 vs. negative control group. MCF-7/s, wild-type drug-sensitive MCF-7 breast cancer cells; DRβ-H, DRhamnose β-hederin; miR, micro RNA.
Target prediction and pathway analysis of miRNAs
To evaluate the biological processes regulated by miR-130a and miR-425, their targets were investigated using TargetScan and miRDB. Only the genes listed by these two independent algorithms were chosen for further study. A total of 443 genes were detected and a KEGG analysis was performed using the DAVID program. As presented in Table I, the predicted genes were suggested to participate in several signaling pathways, including mammalian target of rapamycin (mTOR), ErbB, mitogen activated protein kinase (MAPK) and transforming growth factor (TGF)-β signaling pathways. Furthermore, a number of pathways associated with the tumor metabolic process were also detected, in particular choline metabolism and proteoglycansin cancer.
Table I.
KEGG pathway analysis with DAVID tool.
| Term | Gene count | Percentage | P-value |
|---|---|---|---|
| Protein processing in endoplasmic reticulum | 12 | 2.8 | 3.5×10−3 |
| ErbB signaling pathway | 8 | 1.8 | 6.1×10−3 |
| MAPK signaling pathway | 14 | 3.2 | 1.2×10−2 |
| Choline metabolism in cancer | 8 | 1.8 | 1.3×10−2 |
| TNF signaling pathway | 8 | 1.8 | 1.7×10−2 |
| TGF-β signaling pathway | 7 | 1.6 | 1.9×10−2 |
| Adipocytokine signaling pathway | 6 | 1.4 | 3.1×10−2 |
| AMPK signaling pathway | 8 | 1.8 | 3.4×10−2 |
| Phagosome | 9 | 2.1 | 3.9×10−2 |
| Oxytocin signaling pathway | 9 | 2.1 | 4.6×10−2 |
| Osteoclast differentiation | 8 | 1.8 | 4.7×10−2 |
| mTOR signaling pathway | 5 | 1.1 | 5.8×10−2 |
| Insulin signaling pathway | 8 | 1.8 | 5.9×10−2 |
| Signaling pathways regulating pluripotency of stem cells | 8 | 1.8 | 6.3×10−2 |
| Proteoglycans in cancer | 10 | 2.3 | 6.5×10−2 |
| ECM-receptor interaction | 6 | 1.4 | 6.8×10−2 |
| Calcium signaling pathway | 9 | 2.1 | 8.2×10−2 |
| Circadian entrainment | 6 | 1.4 | 9.1×10−2 |
| Fc epsilon RI signaling pathway | 5 | 1.1 | 9.2×10−2 |
Discussion
A large number of active components purified from natural plants have been used to treatvarious tumors including breast cancer (2,3). The authors' previous study demonstrated that DRβ-H, a noveloleanane-type triterpenoid saponin derived from Clematis ganpiniana, exhibited apotentinhibitory effecton breastcancer cells (5). In the present study, it was reported that DRβ-H may inhibit breast cancer cell growth by remodeling the tumor microenvironment via reducing exosome release.
DRβ-Hwasable to reduce proliferation and induce apoptosis of breast cancer cells. This was based on MTT assay results and confirmed with flow cytometry analysis of cancer cells incubated with different concentrations of DRβ-H. It was previously reported that DRβ-H regulated the phosphoinositide 3-kinase/AKT serine threonine kinase signaling pathway and activated pro-apoptotic proteins of the B cell lymphoma-2 family (5). No apoptosis-associated signaling pathways were examined in the present study, and it will be necessary to investigate the concrete pathways involved in proliferation inhibition.
Exosomes are small vesicles between 50 and 100 nm in diameter that are implicated with several key functions during tumor progression, including angiogenesis, immunosuppression, invasion and metastasis (9). Intercellular communication is also one such function, via exosomes' ability to internalize into surrounding cellsand facilitate constant transfer of active miRNAs (10,21). The authors previous study demonstrated that drug-resistant breast cancer cells were able to spread resistance capacity to sensitive cells viaexo somes and that such effects were partially attributed to the cell-to-cell shuttle of specific miRNAs (12,13). In the present study, it was identified that DRβ-H downregulated the production of tumor-derived exosomes. It was further assessed whether exosomes affected the growth of breast cancer cells. The results indicated that exosomes enhanced the proliferation and attenuated apoptosis following absorption and internalization by target breast cancer cells. The observation of exosomes moving from donor cells to the same type of recipient cells opens up an intriguing possibility that malignant capacity maybe transferred via exosomes. However, the exposure of breast cancer cells to DRβ-H may trigger cells to release less exosomes compared with that in normal conditions, leading to reduced cell growth.
By comparing the miRNA microarray of exosomes and their cells of origin (data not shown), it was identified that exosomes contained a series of miRNAs shared with their parental cells. Additionally, several specific miRNAs were present dominantly or at higher levels in exosomes compared with that in the donor cells. These results are consistent with certain other studies, suggesting that loading of miRNAs into exosomes may not be a random event, but instead is regulated by a selective process (22,23). Asit is difficult to determine the functions of all the exosomal miRNAs, the focus of the present study was shifted to explore the roles of the five miRNAs most strongly localized in exosomes in breast cancer cells. In the present study, only exosomal miR-130a and miR-425 significantly increased cell viability. KEGG pathway analysis of the predicted targets of the two miRNAs demonstrated that the target genes are associated with them TOR, ErbB, MAPK and TGF-β signaling pathways. It is notable that all these signaling pathways are important for cell proliferation and survival (24–27).
There are certain limitations of the present study. It has not been demonstrated that exosomal miR-130a and miR-425 were shuttled into recipient cells and serveas functional molecules to participate a certain signaling pathway. Nevertheless, as confirmed by a number of studies, exosomal miRNAs exert gene silencing to fine-tune target expression through the same mechanism as endogenous miRNAs (13,28,29). Future studies may attempt to separately investigate the role of exosomal miRNAs, and assess the biological significance of secreted miR-130a and miR-425 in the regulation of malignant progression.
In conclusion, the present study expands on previous findings and, to the best of our knowledge, provides the first evidence suggesting that DRβ-H may suppress breast cancer cell growth by manipulating the tumor microenvironment via inhibition of exosome release.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural Science Foundation of China (grant nos. 81702591 and 81502294), the Natural Science Foundation of Jiangsu Province (grant no. BK20170294) and Science Foundation of Changzhou (grant no. CJ20159044).
Availability of data and materials
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
Conceived and designed the experiments: WC, LC and QD. WC, LC and MP performed the experiments. WC, LC, QQ, YZ, and LX analyzed the data. WC and LC wrote the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67:439–448. doi: 10.3322/caac.21412. [DOI] [PubMed] [Google Scholar]
- 2.Wang CY, Bai XY, Wang CH. Traditional Chinese medicine: A treasured natural resource of anticancer drug research and development. Am J Chin Med. 2014;42:543–559. doi: 10.1142/S0192415X14500359. [DOI] [PubMed] [Google Scholar]
- 3.Cragg GM, Newman DJ. Natural products: A continuing source of novel drug leads. Biochim Biophys Acta. 2013;1830:3670–3695. doi: 10.1016/j.bbagen.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding Q, Yang LX, Yang HW, Jiang C, Wang YF, Wang S. Cytotoxic and antibacterial triterpenoids derivatives from Clematis ganpiniana. J Ethnopharmacol. 2009;126:382–385. doi: 10.1016/j.jep.2009.09.028. [DOI] [PubMed] [Google Scholar]
- 5.Cheng L, Xia TS, Wang YF, Zhou W, Liang XQ, Xue JQ, Shi L, Wang Y, Ding Q. The apoptotic effect of D Rhamnose β-hederin, a novel oleanane-type triterpenoid saponin on breast cancer cells. PLoS One. 2014;9:e90848. doi: 10.1371/journal.pone.0090848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chin AR, Wang SE. Cancer-derived extracellular vesicles: The ‘soil conditioner’ in breast cancer metastasis? Cancer Metastasis Rev. 2016;35:669–676. doi: 10.1007/s10555-016-9639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valadi H, Ekström K, Bossiös A, Sjostrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 8.Yu X, Odenthal M, Fries JW. Exosomes as miRNA carriers: Formation-function-future. Int J Mol Sci. 2016;17(pii):E2028. doi: 10.3390/ijms17122028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu DD, Wu Y, Shen HY, Lv MM, Chen WX, Zhang XH, Zhong SL, Tang JH, Zhao JH. Exosomes in development, metastasis and drug resistance of breast cancer. Cancer Sci. 2015;106:959–964. doi: 10.1111/cas.12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: A new form of intercellular communication. Trends Cell Biol. 2012;22:125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Zhong S, Li W, Chen Z, Xu J, Zhao J. MiR-222 and miR-29a contribute to the drug-resistance of breast cancer cells. Gene. 2013;531:8–14. doi: 10.1016/j.gene.2013.08.062. [DOI] [PubMed] [Google Scholar]
- 12.Chen WX, Liu XM, Lv MM, Chen L, Zhao JH, Zhong SL, Ji MH, Hu Q, Luo Z, Wu JZ, Tang JH. Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PLoS One. 2014;9:e95240. doi: 10.1371/journal.pone.0095240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen WX, Cai YQ, Lv MM, Chen L, Zhong SL, Ma TF, Zhao JH, Tang JH. Exosomes from docetaxel-resistant breast cancer cells alter chemosensitivity by delivering microRNAs. Tumour Biol. 2014;35:9649–9659. doi: 10.1007/s13277-014-2242-0. [DOI] [PubMed] [Google Scholar]
- 14.Miot S, Gianni-Barrera R, Pelttari K, Acharya C, Mainil-Varlet P, Juelke H, Jaquiery C, Candrian C, Barbero A, Martin I. In vitro and in vivo validation of human and goat chondrocyte labeling by green fluorescent protein lentivirus transduction. Tissue Eng Part C Methods. 2010;16:11–21. doi: 10.1089/ten.tec.2008.0698. [DOI] [PubMed] [Google Scholar]
- 15.Corcoran C, Rani S, O'Brien K, O'Neill A, Prencipe M, Sheikh R, Webb G, McDermott R, Watson W, Crown J, O'Driscoll L. Docetaxel-resistance in prostate cancer: Evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PLoS One. 2012;7:e50999. doi: 10.1371/journal.pone.0050999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaiswal R, Luk F, Gong J, Mathys JM, Grau GE, Bebawy M. Microparticle conferred microRNA profiles-implications in the transfer and dominance of cancer traits. Mol Cancer. 2012;11:37. doi: 10.1186/1476-4598-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 18.Wang X. Computational prediction of microRNA targets. Methods Mol Biol. 2010;667:283–295. doi: 10.1007/978-1-60761-811-9_19. [DOI] [PubMed] [Google Scholar]
- 19.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(Database issue):D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID bioinformatics resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server Issue):W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen WX, Zhong SL, Ji MH, Pan M, Hu Q, Lv MM, Luo Z, Zhao JH, Tang JH. MicroRNAs delivered by extracellular vesicles: An emerging resistance mechanism for breast cancer. Tumour Biol. 2014;35:2883–2892. doi: 10.1007/s13277-013-1417-4. [DOI] [PubMed] [Google Scholar]
- 22.Palma J, Yaddanapudi SC, Pigati L, Havens MA, Jeong S, Weiner GA, Weimer KM, Stern B, Hastings ML, Duelli DM. MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res. 2012;40:9125–9138. doi: 10.1093/nar/gks656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 24.Dey N, De P, Leyland-Jones B. PI3K-AKT-mTOR inhibitors in breast cancers: From tumor cell signaling to clinical trials. Pharmacol Ther. 2017;175:91–106. doi: 10.1016/j.pharmthera.2017.02.037. [DOI] [PubMed] [Google Scholar]
- 25.Zhou BP, Hung MC. Dysregulation of cellular signaling by HER2/neu in breast cancer. Semin Oncol. 2003;30:38–48. doi: 10.1053/j.seminoncol.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Shao N, Ding X, Tan B, Song Q, Wang N, Jia Y, Ling H, Cheng Y. Crosstalk between transforming growth factor-beta signaling pathway and long non-coding RNAs in cancer. Cancer Lett. 2016;370:296–301. doi: 10.1016/j.canlet.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Miyamoto Y, Suyama K, Baba H. Recent advances in targeting the EGFR signaling pathway for the treatment of metastatic colorectal cancer. Int J Mol Sci. 2017;18(pii):E752. doi: 10.3390/ijms18040752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem. 2013;288:10849–10859. doi: 10.1074/jbc.M112.446831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morel L, Regan M, Higashimori H, Ng SK, Esau C, Vidensky S, Rothstein J, Yang Y. Neuronal exosomal miRNA-dependent translational regulation of astroglial glutamate transporter GLT1. J Biol Chem. 2013;288:7105–7116. doi: 10.1074/jbc.M112.410944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.