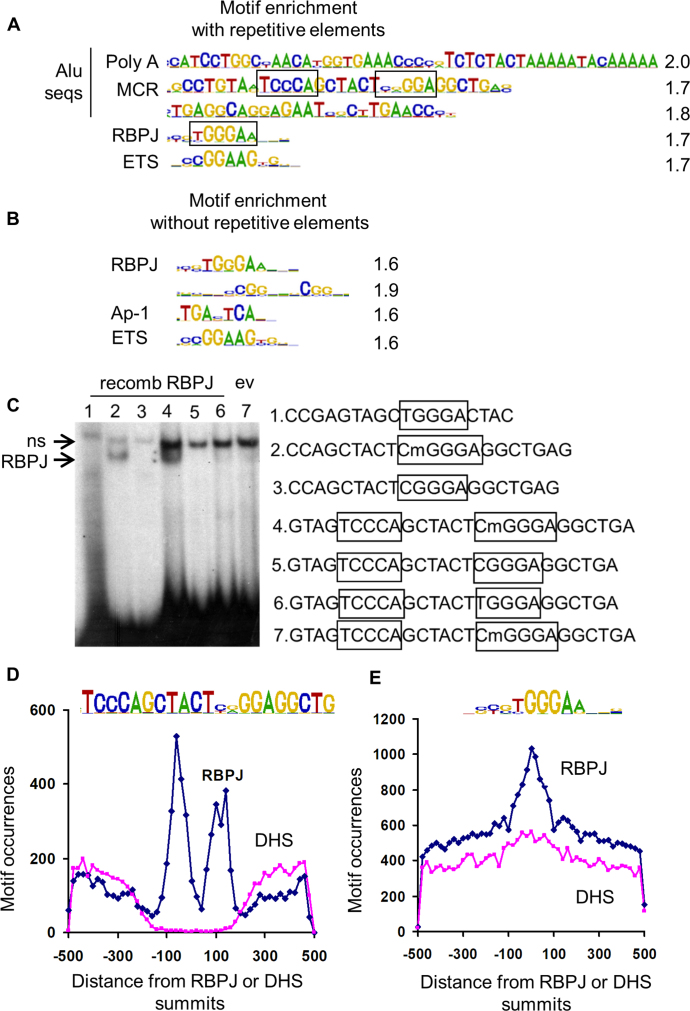
Figure 1.
ChIP-seq analysis of RBPJ binding sites in HuAo SMC identified consensus and methylated Alu motifs. Motif over-representation at RBPJ ChIP-seq peaks summits (±100 bp) when repetitive elements were included (A) or not included (B) in the analysis. Quantification of motif enrichment is listed at right, and putative RBPJ binding sites are boxed. (C) Gel shift assays were performed by incubating RBPJ-containing COS cell lysates with the methylated or unmethylated double stranded DNA probes listed at right. Note that RBPJ interacted only with probes containing the CmGGGA sequence (lower arrow). Note that the binding reaction in lane 7 contained lysates from COS cells transfected with empty expression vector, and the upper arrow marks a non-specific, non-RBPJ-containing binding complex. (D) Distribution of Methylated Cytosine RBPJ binding elements near RBPJ peak summits or 30 000 random DNase hypersensitivity (DHS) regions summits. (E) Distribution of consensus RBPJ binding elements near RBPJ peak summits or 30 000 random DHS regions summits.