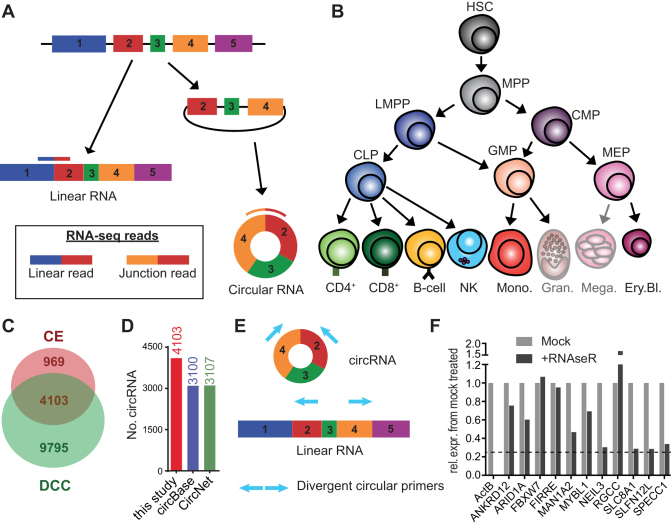
Figure 1.
Circular RNA expression in hematopoietic cells. (A) Diagram presenting the forward-splicing of a linear RNA and the back-splicing of a circular RNA. (B) Diagram of hematopoietic differentiation (inspired by ref (27)). HSC: Hematopoietic stem cell, MPP: Multipotent progenitor, LMPP: lymphoid-primed multipotent progenitor, CLP: common lymphoid progenitor, CMP: common myeloid progenitor, GMP: Granulocyte-macrophage progenitor, MEP: Megakaryocyte-erythrocyte progenitor, CD4: CD4+ T cells, CD8: CD8+ T cells, NK: Natural killer cells, Mono.: Monocytes, Gran.: Granulocytes, Mega.: Megakaryocytes, Ery.Bl.: Erythroblasts. (C) The number of circRNA (≥2 junction reads in at least one sample of a specific cell population) detected in hematopoietic cells using DCC and CircExplorer2 (CE). (D) Numbers of circRNA detected by DCC and CE combined using low confidence cut-off, compared to circRNA annotated in circBase and in CircNet. CircRNA that were not previously annotated were 1003 for circBase and 996 for CircNet. (E) Scheme of primer design for detection by PCR of circular RNA. (F) Graph showing the remaining expression after RNAseR treatment relative to mock treated for different circRNA. β-actin mRNA was used as a linear transcript control. Dashed line depicts the 50-fold higher resistance to RNAseR treatment, compared to the β-actin linear control (as in (6)). Data are representative of 2 donors.