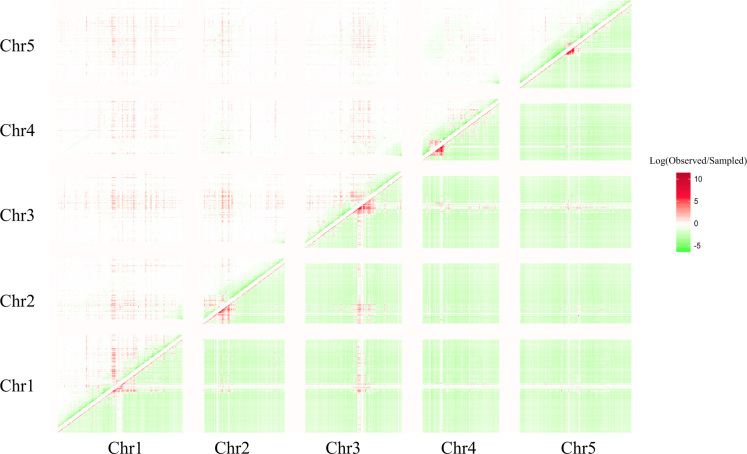
Figure 3.
Heatmap of the natural logarithm of the ratio of the number of observed pair hits, Np, (correlated SNP pairs; counts per bins of 100K bp) and (A) lower-right triangular matrix: the number of pairs estimated purely based on the density of SNPs, and (B) upper-left triangular matrix: the number of random pairs forced to follow the same distance distribution (Figure 2). In (A), the number, Np,r, of density-derived, random pairs per cell (i, j) (bin i (x-axis), bin j (y-axis) was obtained by Np,r = Ni*Nj/T, where Ni,j are the number of SNPs in bin i, j, respectively, and T is a scaling factor to render to the total number of actual and density-based pairs identical. To both, the numerator and the denominator, an arbitrary constant of 10 was added to avoid dividing by zero and to mitigate the noise due to small numbers. Note that, also in the case of distance-biased re-sampling, the emerging clusters of elevated pair hit frequency do not reflect local SNP-density as the random re-sampling does consider local SNP density (see Methods).