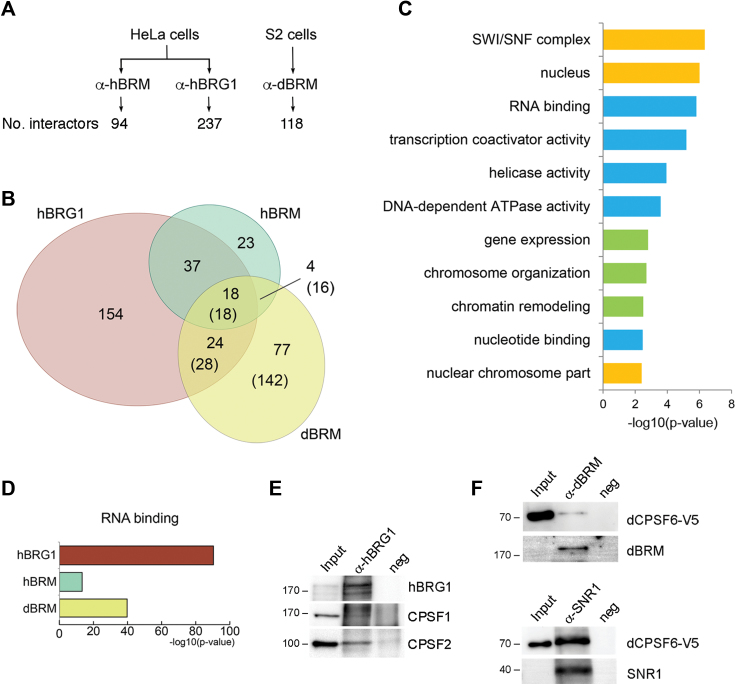
Figure 7.
SWI/SNF interacts with pre-mRNA processing factors in fly and human cells. Endogenous dBRM, hBRM and hBRG1 were immunoprecipitated from S2 and HeLa cells, respectively, and the immunoprecipitated proteins were identified by mass spectrometry. (A) The figure shows the number of interactors identified for each of the SWI/SNF ATPases by IP and label-free mass spectrometry. (B) Euler diagram showing the overlaps among the proteins coimmunoprecipitated with each ATPase. The diagram includes only hits with annotated orthologs in D. melanogaster. For D. melanogaster proteins, the number of human orthologues is given in parentheses. (C, D) PANTHER overrepresentation tests using the complete GO database. The p values shown in the figures were calculated using a Fisher's exact test with FDR correction and are expressed –log10(P-value). (C) Selected GO terms enriched in the group of 18 proteins from the intersection between hBRG1, hBRM and dBRM human orthologs. Biological processes, cellular compartments and molecular functions are shown in green, yellow and blue, respectively. (D) Overrepresentation of the GO term ‘RNA binding’ in the interactomes of hBRG1, hBRM and dBRM human orthologs. (E) Western blot analysis of coimmunoprecipitated proteins showing the interaction of human CPSF1 and CPSF2 with hBRG1. (F) Endogenous dBRM or SNR1 were immunoprecipitated from S2 cells that expressed V5-tagged CPSF6. CPSF6 is interacts with dBRM and SNR1. The approximate mobility of molecular mass standards is shown to the left of the blots, in kDa.