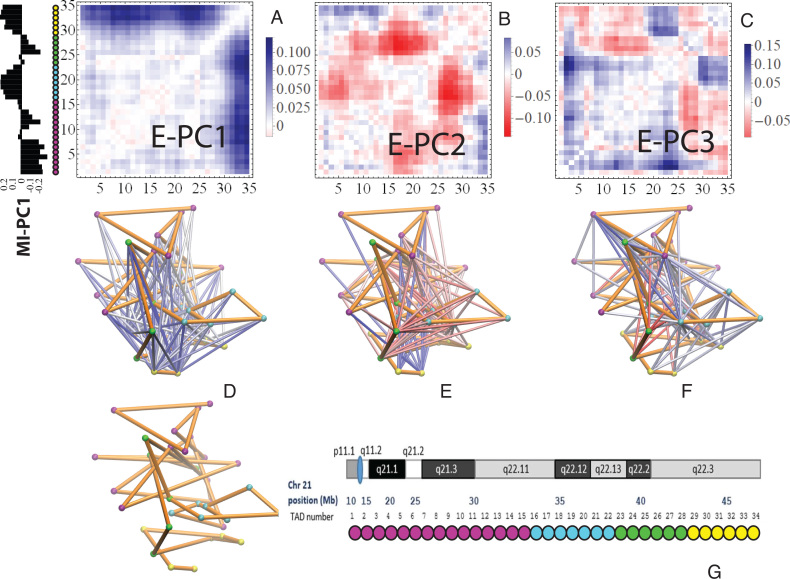
Figure 3.
E-PCA applied to chr21 TAD imaging data reveals modes of chromosome fluctuation across individual IMR90 cells. The displacement matrix view of PC1, PC2, PC3 of E-PCA are shown in (A–C). PC1 from MI-PCA on the same system is shown as a bar-graph at left and TAD elements are colored according to general domains detected by E-PCA. The corresponding 3D representations are shown in (D–F), where the values of displacement matrix elements 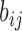 are used to color cylinders that connect TADs
are used to color cylinders that connect TADs 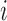 and
and 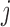 . Less significant relationships (cutoff is
. Less significant relationships (cutoff is 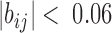 ) are not shown. The reference 3D path of the 34 TADs (the configuration detected for a chosen cell, cell #1) is shown in (G) along with a reference for the genomic position of each TAD probe. Explanation was made in SI for the selection of cell #1 as the reference.
) are not shown. The reference 3D path of the 34 TADs (the configuration detected for a chosen cell, cell #1) is shown in (G) along with a reference for the genomic position of each TAD probe. Explanation was made in SI for the selection of cell #1 as the reference.