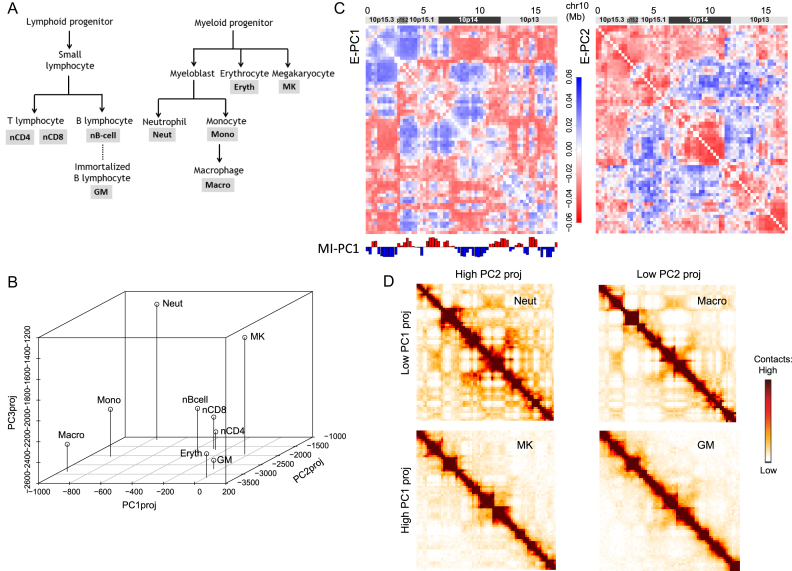
Figure 4.
E-PCA applied to Hi-C data from 9 different blood cell types provides insight about chromosome structure differences between cell types. (A) The blood cell lineage from which the Hi-C data are derived, showing developmental relationships. Abbreviations used in subsequent panels are defined. (B) The projection of each cell type's Hi-C contact map onto E-PC1, 2 and 3. Cell types segregate in this space according to lineage relationships. (C) The displacement matrix for E-PC1 and E-PC2, showing the 17 Mb region of chromosome 10 from which the sample Hi-C data was drawn. (D) Representative original input Hi-C contact matrices for this same region of chr10 for cell types with high and low E-PC1 or PC2 projections, visibly showing the contact pattern differences between these categories of cell types.