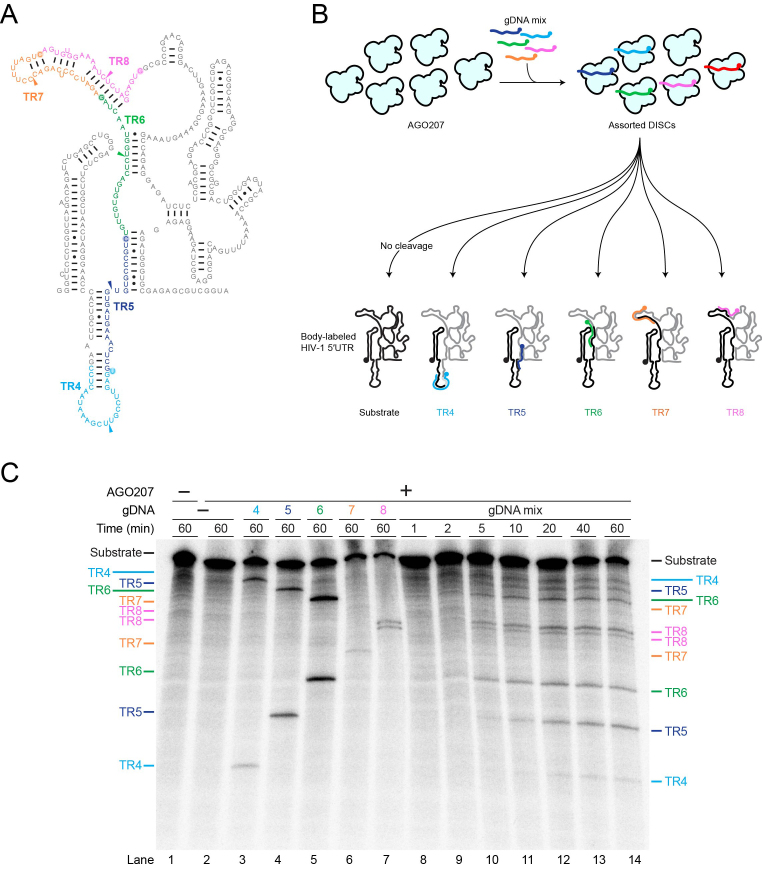
Figure 5.
Detection of multiple cleavage events in a single reaction. (A) Secondary structure of HIV-1 ΔDIS 5′UTR with TRs colored for clarity. Colored arrowheads indicate cleavage sites on each TR. (B) Schematic of cleavage assay using multiple gDNAs targeting different TRs of HIV-1 ΔDIS 5′UTR. For clarity, AGO207 molecules bound to endogenous E. coli RNA are not shown. AGO207 was loaded with five gDNAs (gDNA4–gDNA8). Following assembly of the DISCs, a 32P body-labeled HIV-1 ΔDIS 5′UTR RNA was added to the mixture to initiate cleavage. Cartoon secondary structure showing cleavage products with the 5′ products colored black and 3′ products colored gray. Colored gDNAs show complementary regions between guide and target strands. (C) Multiple DISC-mediated cleavage. Substrates and products formed at the indicated time points were separated by denaturing PAGE (8%). Reactions using only one guide are shown in lanes 3–7 as reference for product migration. The one-pot reaction time course is shown in lanes 8–14.