Abstract
The catalytic DNA circuits play a critical role in engineered biological systems and molecular information processing. Actually, some of the natural or synthetic DNA circuits were triggered by covalent modifications, where conformational changes were induced to facilitate complex DNA engineering functions and signal transmissions. However, most of the reported artificial catalytic DNA circuits were regulated by the toehold-mediated reaction. Therefore, it is significant to propose a strategy to regulate the catalytic DNA circuit not only by the toehold-mediated mechanism, but also by involving the conformational changes induced by the covalent modification. In this study, we developed the catalytic DNA logic circuits regulated by DNAzyme. Here, a regulation strategy based on the covalent modification was proposed to control the DNA circuit, combing two reaction mechanisms: DNAzyme digestion and entropy-driven strand displacement. The DNAzyme and DNA catalyst can participate into the reactions alternatively, thus realizing the cascading catalytic circuits. Using the DNAzyme regulation, a series of logic gates (YES, OR and AND) were constructed. In addition, a two-layer cascading circuit and a feedback self-catalysis circuit were also established. The proposed DNAzyme-regulated strategy shows great potentials as a reliable and feasible method for constructing more complex catalytic DNA circuits.
INTRODUCTION
DNA is a promising material in the field of nanotechnology, owing to the predictable Watson–Crick base pairing, the outstanding data-storage capacity and tininess (1). To date, DNA has been exploited to construct logic operations (2–7), cascading networks (8–13) and cycling circuits (14–19), demonstrating its great potential in molecular nano-engineering and computing. In particular, DNA logic circuit plays a critical role in engineered biological systems as regulation of signal amplification and information processing. Accordingly, the functions of signal delivery and propagation are performed via the various DNA circuits. In the DNA-based signaling networks, most of signaling operations employed two main mechanisms (20–27): non-catalytic and entropy-driven catalytic DNA circuits. In catalytic DNA circuits, the input DNA trigger can be recycled for multiple rounds and maintains itself without being consumed. Therefore, the catalytic DNA mechanism provides more possibilities to perform cascading information process, such as signal amplification and transmission (25–27).
The main regulation method in the catalytic DNA circuits is toehold-mediated strand displacement that was simple, robust and modular and widely used to trigger the circuits and connect up and downstreams (28,29). In most of the toehold-regulated catalytic circuits, the rearrangements of DNA hybridization occurred during strand displacement reactions, without any covalent modification (28,29). In fact, in some of the natural or synthetic DNA circuits, the reactions were triggered by covalent modifications (e.g. cleaving backbone bonds) to facilitate complex DNA engineering functions and signal transmissions, such as treated with light, protein enzymes and DNAzymes (30–32). The regulations of the conformational changes can provide more possibilities to control the DNA networks in a modular and tunable manner (33,34). However, most of the reported artificial catalytic DNA circuits were regulated by the toehold-mediated reaction. Therefore, it is significant to propose a strategy to regulate the catalytic DNA circuit not only by the toehold-mediated mechanism, but also involving the conformational changes induced by the covalent modification.
In reality, catalytic nucleic acids (DNAzymes or ribozymes) have been used as active elements for the signal amplification and information delivery to detect small stimuli (e.g. Mg2+, Cu2+ and aptamers) (35–42). Meanwhile, DNAzyme also has been proven to be particularly suitable for DNA logic operations with good regulation and flexible design (43–48), due to the simple synthetic preparation and highly efficient digestion. Briefly, typical DNAzyme digestion can lead to the bond breaks of the DNA substrate to induce the conformational changes, which can release the product strand for the subsequent trigger events. Therefore, such DNAzyme-mediated operation provides an effective route to achieve a well-regulated covalent modification. To the best of our knowledge, despite the well development of the two methods: catalytic DNA circuit and DNAzyme, it has rarely been reported a cascading DNA networks combined with the two components.
Herein, we developed a system of cascading entropy-driven DNA logic gates regulated by DNAzymes. In this system, a regulation strategy based on the covalent modification was proposed to control the DNA circuit, combing two reaction mechanisms: DNAzyme digestion and entropy-driven strand displacement. First, the circuit responds to the conformational changes induced by specific DNAzyme digestion. Subsequently, the entropy-driven favorability is facilitated via strand displacement, to perform a catalytic strand displacement reaction. In the process, the strand displacement reaction leads to the generation of a new DNAzyme, which can then act as an input to trigger the next downstream targets. In our design, DNAzyme and DNA catalyst participated in the reactions alternatively, thus realizing a cascading catalytic circuit. Importantly, DNAzyme in this study plays two regulation roles as both an input trigger and an output signal, which can specifically connect up and downstreams. The DNA catalyst here serves as an unconsumed input trigger. To demonstrate the feasibility and scalability of the DNAzyme-based regulation strategy, a series of ‘logic gates’ (YES, OR and AND) are constructed. Moreover, a two-layer cascading circuit and a feedback self-catalysis logic circuit were also established. The results were confirmed via native polyacrylamide gel electrophoresis (PAGE) and fluorescence detection. The proposed DNAzyme-regulated strategy shows a great potential as a reliable and feasible method for constructing complex cascading DNA circuits, which may have more applications in molecular sensing, nano-device and DNA computing.
MATERIALS AND METHODS
Materials
All the DNA strands were synthesized by Sangon Biotech. Co., Ltd. Unmodified DNA strands were purified by PAGE and modified DNA strands with fluorophore were purified by high-performance liquid chromatography. All DNA strands were dissolved in water as stock solution and quantified using a Nanodrop 2000, and absorption intensities were recorded at λ = 260 nm. The sequences of all strands are listed in Supplementary Table S1. Gels were prepared using TAE/Mg2+ buffer (40 mM Tris, 20 mM acetic acid, 2 mM ethylenediaminetetraacetic acid (EDTA) and 12.5 mM magnesium acetate, pH 8.0) and 500 ml of mother liquor of acrylamide at a concentration of 45% (217 g of acrylamide and 8 g of N,N′-methylenebis (acrylamide)). Other chemicals were of reagent grade and were used without further purification.
Logic gate preparation
Logic gates were formed by mixing several kinds of DNA strands in a course of slow annealing. Various equimolar DNA strands were added to a final concentration of 0.6 μM, in 1× TAE/ Mg2+ buffer (20 mM Tris acetate, 2 mM EDTA, 12.5 mM Mg acetate, pH 8.0). The mixture is annealed at the reaction condition of 95°C for 5 min, 65°C for 30 min, 50°C for 30 min, 37°C for 30 min, 22°C for 30 min and finally kept at 4°C. Subsequently, the catalyst inputs (strands B–45, B1–45 and B2–45) and the input DNAzyme 2 or 3 were added to a final concentration of 0.3 and 1.2 μM and reacted at the room temperature for a night. In a two-layer ‘YES’ logic gate, after first constructing each gate separately, the two-layer ‘YES’ logic gate was then formed by mixing the solutions of all gates.
Native polyacrylamide gel electrophoresis (PAGE)
The DNA solutions mixed with 6× loading buffer (Takara) were analyzed in a 12% native PAGE was conducted in 1× TAE/Mg2+ buffer at a constant voltage of 100 V for 2 h.
Fluorescent biosensing assays
In the fluorescent assays of all logic gates (YES, OR, AND and two-layer YES), 0.4 μM of the gate strands solution was incubated with the catalyst inputs (0.1–0.2 μM), and input DNAzyme (1.2 μM) in 1× TAE/Mg2+ buffer, and fluorescence intensities were recorded every 6 min. All fluorescence experiments were repeated three times to ensure reproducibility. The fluorescent experiments were implemented using real-time polymerase chainreaction (PCR) (Agilent, Mx3005P).
RESULTS AND DISCUSSION
Basic YES gate controlled by strand displacement
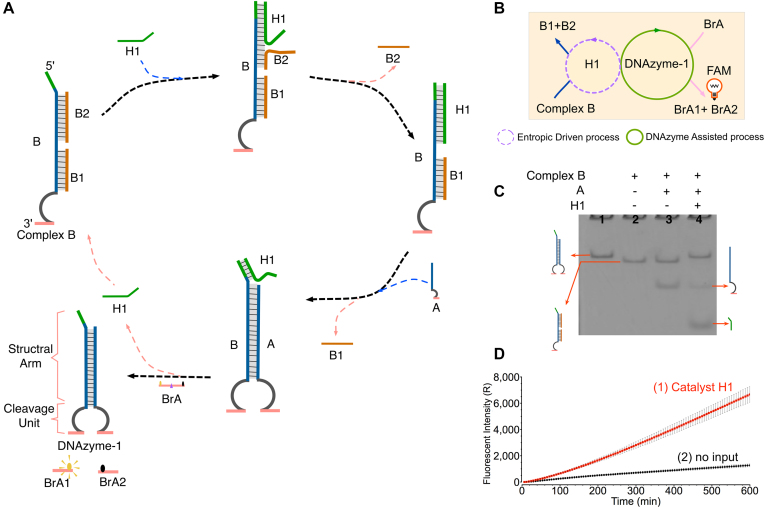
As shown in Figure 1A, the ‘YES’ gate is triggered by a DNA strand to produce DNAzyme (DNA →DNAzyme). The DNAzyme was designed to be separated into two individual parts: strands A and B. Strand B was initially protected by embedding into a DNA complex B (B/B1/B2). Only when A and B hybridized, the complete DNAzyme can be produced to serve as an active output reporter to produce fluorescence signals by cutting substrate BrA. Here, strand BrA was designed to have a ribonucleotide cleavage site (TrAGG) in the middle region with the fluorophore carboxy-fluorescein (FAM) and quencher BHQ functionalized at either end. The DNAzyme cutting can result in the separation between fluorophore and quencher, thus causing increasing fluorescence intensity. In this gate, two catalytic processes are employed as entropy-driven and DNAzyme catalysis mechanisms. Accordingly, the structure of DNAyzme was designed as two functional parts: a structural arm and a cleavage unit (Figure 1A). Notably, entropy-driven DNA strand displacement was used to control the hybridization state of the structural arm. Complex B and strand A initially coexist in solution without triggering. The reaction can only be triggered upon addition of catalyst H1. Specifically, catalyst H1 can first hybridize with the DNA complex through a 6-nt toehold, thus resulting in disassociation of B2 from the complex. Then, a 4-nt newly exposed single-stranded region is generated on strand B to facilitate downstream strand displacements. Subsequently, strand A can hybridize with strand B via the 4-nt toehold to form an active DNAzyme-1, simultaneously releasing the preoccupied B1 and catalyst H1. In this case, catalyst H1 can be recycled for multiple times to participate the reaction. At last, the formed DNAzyme-1 can digest the fluorescent substrate BrA resulting in a significant fluorescence increase. Here, the reaction can be depicted as an abstract graph in Figure 1B, where the dotted circle and the solid circle represent the entropy-driven catalysis and DNAzyme catalysis, respectively.
Figure 1.
(A) Scheme for the ‘YES’ gate using the DNAzyme-based regulating strategy. (B) The abstract graph of the YES gate. The purple and green circles represent the two catalysis amplifications as entropy-driven and enzymatic mechanisms, respectively. (C) Gel analysis of the YES gate reaction using a 12% PAGE. Lane 1, DNAzyme 1 consisting of strands B and A; lane 2, the previous complex B; lane 3, the previous B complex with addition of strand A; lane 4, the B complex and A of the gate strands after adding catalyst H1. (D) Time-dependent fluorescent intensity changes in the presence of (1) input strand H1 or (2) no input. The time interval is 6 min. All data represent the average of three replicates. Error bars represent one standard deviation from triplicate analyses.
The reaction of YES gate was confirmed by native PAGE gel electrophoresis and fluorescence assay (Figure 1C and D). As shown in Figure 1C, lane 3, no displacement reaction occurred without addition of catalyst H1 and thus the two individual bands corresponding to the B complex and A can be observed. However, in the presence of catalyst H1, the gel band of B complex disassembled to produce a new band of DNAzyme-1 (lane 4). In addition, to test the signal amplification ability of catalyst H1, a control experiment was conducted by reducing the concentration of catalyst H1 to trigger the reaction (Supplementary Figure S1a). The results showed that a significant production of DNAzyme-1 can be found even at a low ratio of [H1]: [DNA complex] = 1:6, thus indicating the successful catalytic ability of H1.
In addition, a fluorescence assay was conducted to monitor the ‘YES’ gate in real time. A significant increase in fluorescence signals was observed upon addition of the catalyst H1 (Figure 1D, curve 1), confirming that the DNAzyme-based regulating reaction had indeed occurred. To determine the appropriate concentration of catalyst H1 to trigger the reaction, control experiments were also performed using a series of different concentrations of the catalyst. As shown in Supplementary Figure S1b, with the increases of H1 concentrations, the fluorescent intensity increased correspondingly. Based on the results above, 0.1–0.2 μM was chosen as the appropriate concentration of catalyst H1 in different logic operations. Notably, the fluorescence signal increased at a slow rate in curve 1. We attribute this phenomenon as the low concentration of the gate strands leads to an incomplete consumption of the fluorescence substrate BrA during 10 h reaction time. In the Supplementary Data, Supplementary Figure S1c, a control experiment was implemented by increasing the gate strands concentrations to completely consume BrA and extending the reaction to 16 h. Accordingly, the fluorescence signals showed rather fast growths at early time points and quickly reached the top level.
DNAzyme triggered cascading YES gate
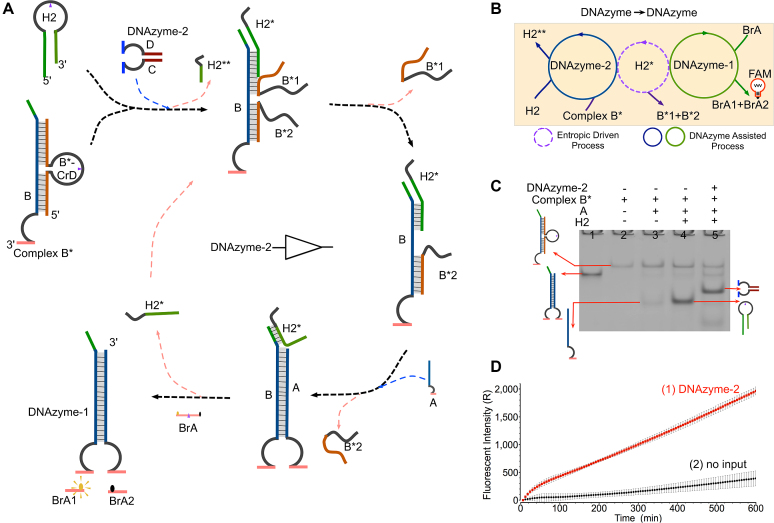
To investigate the feasibility of constructing the hierarchical DNAzyme-based regulating circuit, a cascading YES gate was also developed, in which the circuit was designed to be triggered by the DNAzyme input and generated another DNAzyme. In this system, DNAzyme-2 was designed as the input molecule to trigger the cascading YES gate and produce DNAzyme-1 to induce fluorescence intensity increase. There were four DNA components as DNA complex B* (the left half of DNAzyme protected by strand B*-CrD), strand A (the right half of DNAzyme), the hairpin pre-catalyst H2 and the fluorescent substrate BrA (Figure 2A). Notably, both strand B*-CrD and the pre-catalyst H2 were designed to contain a TrAGG in the middle of the loop region. Therefore, upon addition of DNAzyme-2, the cleavage site in strand B*-CrD will be cut into two pieces, leading to a conformational change to activate DNA complex B*. Meanwhile, DNAzyme-2 also cuts the loop of hairpin H2 to release catalyst H2*, which can initiate the entropy-driven strand displacement and result in the generation of DNAzyme-1. Similar to the basic YES gate described above, catalyst H2* can be recycled multiple times to participate in the reaction toward generating DNAzyme-1. At last, the generated DNAzyme-1 can directly digest the fluorescent substrate BrA to induce a significant fluorescence increase. The reaction can be depicted as an abstract graph (Figure 2B), where the dotted circle, the solid circles represent the catalysis of H2*, DNAzyme-2 and DNAzyme-1, respectively.
Figure 2.
A cascading ‘YES’ logic gate based on the DNAzyme-based regulating strategy. (A) Illustration of a cascading YES gate. The loop region of strand B*-CrD has a TrAGG (purple arrow), which is similar to the pre-catalysts H2. The fluorophore FAM and the quencher BHQ are functionalized at either end of strand BrA. DNAzyme-1 can cleave the reporter strand BrA to trigger fluorescent signals. (B) The abstract graph of two enzymatic catalytic processes (DNAzyme-2 and DNAzyme-1) and one entropy-driven amplification process (catalyst strand H2*) in the cascading ‘YES’ logic reaction. (C) Analysis of YES-gate products using a 12% PAGE gel: ‘+’ denotes addition of the strand and ‘−’ denotes absence of the strand. Lane 1, the products of DNAzyme-1; lane 2, the product of the duplex substrate B*; lane 3, the gate stands consist of strand A and the duplex substrate; lane 4, the gate strands and one hairpin pre-catalyst H2; lane 5, the product after adding DNAzyme-2. (D) Time-dependent fluorescence changes according to different inputs. The time interval is 6 min. Curve (1) reflects the reaction with the addition of DNAzyme-2 and curve (2) is the case with no input. All data represent the average of three replicates. Error bars represent one standard deviation from triplicate analyses.
To verify the cascading YES-gate operation, both a fluorescence assay and PAGE experiments were carried out. A significant fluorescence signal increase was observed in the presence of DNAzyme-2 (Figure 2D, curve 1), whereas no such increase was detected in the absence of input DNAzyme-2 (Figure 2D, curve 2). Similarly, analysis of the gel banding patterns showed that the addition of DNAzyme-2 resulted in a newly formed band corresponding to DNAzyme-1 along with digestion of the hairpin pre-catalyst H2 (Figure 2C, lane 5). However, when DNAzyme-2 was absent, no product of DNAzyme-1 was generated and the pre-catalyst H2 remained intact (Figure 2C, lane 4). To determine the most suitable reaction concentration, a control PAGE gel and fluorescence experiments were conducted with varying concentrations of DNAzyme-2 (0.03–3 μM) (Supplementary Figure S2).
Two-layer cascading YES gate controlled by DNAzyme
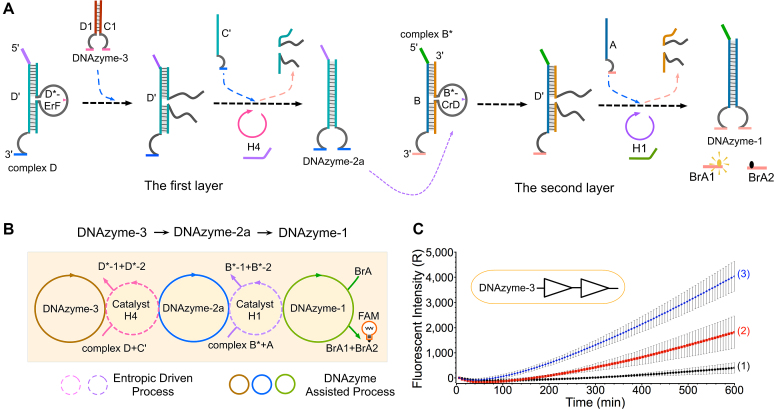
To test the expandability of HDCD-based circuit, we also established a two-layer cascading YES gate regulated as DNAzyme-3→DNAzyme-2a→DNAzyme-1. As illustrated in Figure 3A, DNAzyme-3 was designed as the input in the first-layer YES gate to trigger the activation of complex D (targeting the loop cleavage site of strand D*-ErF). Then, with the help of catalyst H4, the complementary strand C’ can displace strands D*-1 and D*-2 via the entropy-driven mechanism, thus generating complete DNAzyme-2a. Subsequently, the newly formed DNAzyme-2a serves as the input to trigger the second-layer YES gate. Similarly, triggered by DNAzyme-2a, the state of DNA complex B* in the second-layer gate can be activated. Assisted by catalyst H1, DNAzyme-1 can be generated to induce an increasingly fluorescence intensity by digesting reporter BrA. The reaction can be depicted as an abstract graph in Figure 3B, where the dotted circles represent the catalysis of H4 and H1, the solid circles represent the catalysis of DNAzyme-3, DNAyzme-2a and DNAzyme-1, respectively.
Figure 3.
(A) Illustration of a two-level cascading circuit YES gate. The first-level YES gate consists of the duplex complex D, single strand C’, catalyst H4 and input DNAzyme-3. The second-level YES gate is composed of the duplex complex B*, single strand A, fluorescent reporter BrA and catalyst H1. The output DNAzyme-2a in the first-level gate triggers the downstream reaction as an input to induce a significant fluorescence increase. (B) Illustration of three DNAzyme catalytic reactions (DNAzyme-3, DNAzyme-2a and DNAzyme-1) and two entropy-driven reactions (catalysts H4 and H1) in the two-level cascading circuit. (C) Time-dependent fluorescence changes with different inputs. The time interval is 6 min. Curves (1), (2) and (3) demonstrate the effect of the addition of (1) DNAzyme-3 without catalysts H1 and H4, (2) with catalysts H1 and H4 in the absence of DNAzyme-3, (3) in the presence of DNAzyme-3 and both catalysts, respectively. All data represent the average of three replicates. Error bars represent one standard deviation from triplicate analyses.
In the fluorescence assay, a significant fluorescence output was obtained in the presence of the input DNAzyme-3 and the two catalysts H4 and H1 (Figure 3C, curve 3). However, relatively lower fluorescence leakage was observed in the absence of input DNAzyme-3 (curve 2). This phenomenon possibly attributes to the direct strand invasion of DNA catalysts H4 and H1. To verify this speculation, the control experiments were also implemented in support information Supplementary Figure S3. It was indicated that the leaks were mainly caused by catalyst H1. Meanwhile, no obvious fluorescence signal was observed in the absence of the two catalysts (curve 1). Moreover, PAGE analysis was employed to verify the two-layer cascading YES gate (Supplementary Figure S4), also indicating the successful operation of the two-layer logic DNA circuit. The experimental results were also confirmed via the cascading simulation model in Supplementary Data Section 3 (Supplementary Figure S9).
DNAzyme-triggered OR gate
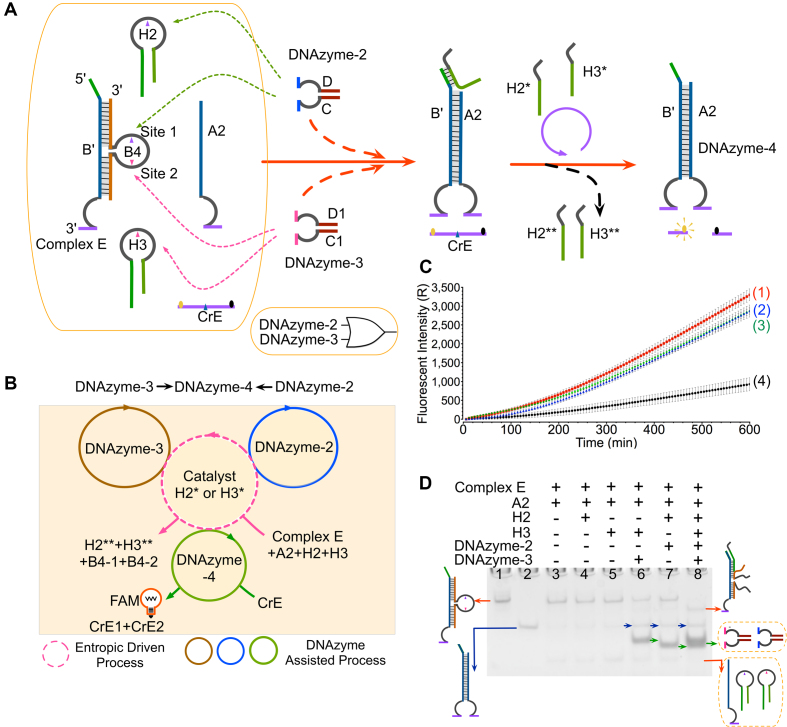
To further test the reliability and expandability of the DNAzyme-based regulating circuit, an ‘OR’ logic gate was constructed triggered by DNAzyme-2 and DNAzyme-3 (DNAzyme-2→DNAzyme-4←DNAzyme-3). DNA components used in this system included complex B’/B4, the fluorescent substrate CrE, catalysts H2 and H3 and the single strand A2 (the right half of DNAzyme-4). In this system, two catalysts, H2 and H3, were employed with a hairpin structure design to reduce possible leakage. Notably, different from the mono-cleavage site design in other gates, the complex E here contains two TrAGGs, corresponding to DNAzyme-2 and DNAzyme-3, respectively (purple arrow and pink arrow in Figure 4A). Meanwhile, two hairpin catalysts H2 and H3 can be activated by digestion of the DNAzyme-2 and DNAzyme-3, respectively. That means when adding any of DNAzyme-2 or DNAzyme-3, the complex E and DNA catalyst can be activated simultaneously via cleavage of loop site, thus resulting in the generation of DNAzyme-4. At last, the formed DNAzyme-4 digests the fluorescent reporter CrE to produce the FAM fluorescence increase. The reaction can be depicted as an abstract graph in Figure 4B, where the dotted circle represents the catalysis of H2 or H3, the solid circles represent the catalysis of DNAzyme-2, DNAyzme-3 and DNAzyme-4, respectively.
Figure 4.
Regulations of the OR gate using DNAzymes-2 and -3. (A) Schematic illustration of the logic gate. The loop region of strand B4 has two TrAGGs (purple arrow, loop site 1; pink arrow, loop site 2), which are similar to catalysts H2 (purple arrow) and H3 (pink arrow), respectively. The fluorophore FAM and quencher BHQ are functionalized at either end of strand CrE. DNAzyme-1 can cleave the reporter strand CrE to trigger fluorescent signals. (B) Abstract graph of three DNAzyme catalytic reactions (DNAzyme-3, -2 and -4) and one entropy-driven reaction (catalysts H2 or H3) in the OR gate. (C) Time-dependent fluorescence changes with different inputs. The time interval is 6 min. The curves (1), (2), (3) and (4) reflect the reactions with the addition of DNAzymes-2 and -3, DNAzyme-2, DNAzyme-3 and no input, respectively. All data represent the average of three replicates. Error bars represent one standard deviation from triplicate analyses. (D) Analysis of OR gate products using a 12% PAGE gel. ‘+’ denotes addition of the strand. Lanes 1, the products of the complex E; lane 2, the product of DNAzyme-4; lane 3, gate stands consisting of strand A2 and the duplex complex; lane 4, gate strands and one hairpin catalyst H2; lane 5, gate strands and one hairpin catalyst H3; lanes 6–8, in the presence of DNAzyme-3, DNAzyme-2 and both, respectively.
In the fluorescence assay, it was observed that in the presence of either DNAzyme-2 or -3, the fluorescence signals increased significantly (Figure 4C, curves 2 and 3). Meanwhile, simultaneous addition of DNAzyme-2 and DNAzyme-3 led to a greater fluorescence increase than those of addition either one trigger input (curve 1). This synergistic effect may arise from the high amount of DNAzyme-4 generated when both triggers were introduced at the same time. In the absence of any input, no significant fluorescence increase can be observed. However, a small amount of fluorescence could still be observed, indicating that a certain degree of leakage occurred in the control reaction (Figure 4C, curve 4).
To better confirm the OR gate operation, a gel electrophoresis experiment was also implemented. In the absence of the two inputs DNAzymes-2 and -3, no gel band representing DNAzyme-4 was observed (Figure 4D, lane 5). Nevertheless, when either DNAzyme-2 or -3 was introduced, a new band of DNAzyme-4 was also generated (blue arrows in lanes 7 and 6) and the gel band corresponding to DNA complex E largely disappeared. Addition of both DNAzymes-2 and -3 also led to the generation of the DNAzyme-4 band (lane 8). Interestingly, the quantities of DNA complex E remaining in the reactions differed according to the input conditions. With the simultaneous addition of two triggers, the band of the DNA complex disappeared without a trace (lane 8), whereas with only one trigger there was still some surplus DNA complex remaining (lanes 6 and 7). These gel results here were nearly identical with the fluorescence results shown in Figure 4C. Similar to the OR gate, an AND gate operation was also constructed, and the results of fluorescence and gel electrophoresis can be found in the Supplementary Figure S5.
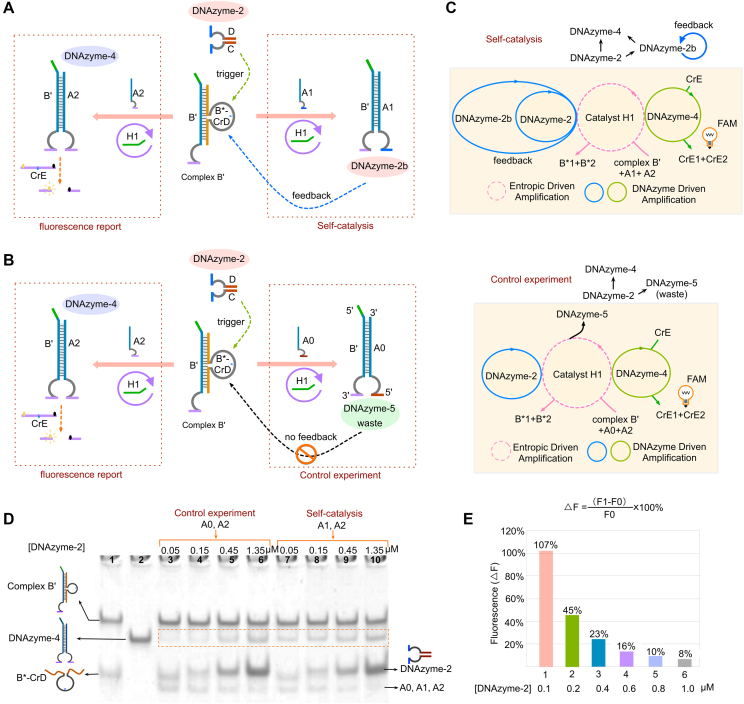
Self-catalysis DNA circuit using feedback mechanism
At last, we designed a self-catalysis DNA circuit, where a feedback loop was established to catalyse the reaction itself (Figure 5A). In this system, the whole circuit was divided into two functional parts: fluorescence reporter and self-catalysis loop, which were simultaneously initiated by DNAzyme-2. After triggering, the DNA complex B’ (B’/B*-CrD) can be activated and the products DNAzyme-2b and -4 were generated in fluorescence reporter and self-catalysis loop, respectively. Because DNAzyme-2b was designed to target at complex B’, as well as initial trigger DNAzyme-2, the feedback loop was constructed as DNAzyme-2→complex B’ →DNAzyme-2b→complex B’ →DNAzyme-2b. Therefore, the feedback circuit can accelerate the generations of both DNAzyme-2b and DNAzyme-4, thus realizing self-catalysis reaction. To compare the self-catalysis effect in detail, a control experiment was also carried out, where the feedback circuit was inhibited via replacing DNAzyme-2b product with the waste product DNAzyme-5 (Figure 5B). In the control experiment, the only initial DNAzyme-2 serves as the trigger and no other self-catalysis trigger produced during the reaction (DNAzyme-2→complex B’→DNAzyme-5). By contrast, in the self-catalysis circuit, the feedback mechanism can accelerate the generation of new trigger DNAzyme-2b, thus significantly promoting the reaction and resulting in higher fluorescence intensity. The reactions can be depicted as abstract graphs in Figure 5C, where the dotted circle represents the catalysis of B3, the solid circles represent the catalysis of DNAzyme-2, DNAyzme-2b and DNAzyme-4.
Figure 5.
(A) Illustration of a self-catalysis circuit. With the help of catalyst H1, DNAzyme-2 is used as an input to trigger the whole circuit, generating two products: DNAzyme-2b and DNAzyme-4. DNAzyme-2b has the same enzyme cleavage site as the input DNAzyme-2 and can therefore cut strand B*-CrD to initiate the self-catalysis reaction. (B) Illustration of a control experiment. DNAzyme-2 is used as an input to trigger the whole circuit, generating two products: DNAzyme-5 and DNAzyme-4. DNAzyme-5 is a waste product. (C) Abstract graph of three DNAzyme catalytic reactions (DNAzyme-2, DNAzyme-2b and DNAzyme-4) and one entropy-driven reaction (catalyst H1) in the self-catalysis circuit and control experiment. (D) Analysis of the self-catalysis circuit products using a 12% PAGE gel. Lanes 1, products of the duplex substrate B*; lane 2, the product of DNAzyme-4; lanes 3–6, a series of different DNAzyme-2 concentrations added to the gate solution with strands A0 and A2 as 0.05, 0.15, 0.45 and 1.35 μM, respectively; lanes 7–10, a series of different DNAzyme-2 concentrations added to the gate solution with strands A1 and A2 as 0.05, 0.15, 0.45 and 1.35 μM, respectively. Other DNA components were used at fixed concentrations 0.6 μM of B’/B*-CrD, 0.15 μM of catalyst H1 and 0.1 μM of ssDNA (A0, A1 and A2). (E) Fluorescence signal analysis. The columns 1–6 correspond to the relative fluorescence increase percentage (△F) before and after self-catalyzing at 0.1, 0.2, 0.4, 0.6, 0.8 and 1.0 μM, respectively.
PAGE analysis was implemented to validate the effects of the self-catalysis feedback (Figure 5D). Because of the feedback effect, a small amount of input DNAzyme-2 was enough to trigger the circuit in the self-catalysis reaction. Therefore, in this experiment, varying concentrations of DNAzyme-2 were used to trigger the reactions as 0.05, 0.15, 0.45 and 1.35 μM. Notably, there was a clear difference in the gel results of the control (lanes 3–6) and self-catalysis (lanes 7–10) experiments. Here, we use the generation quantities of the target products to determine the effects of self-catalysis, including DNAzyme-2b, DNAzyme-5 and DNAzyme-4 (red square frame in Figure 5D). When using 0.05 μM of DNAzyme-2, there was essentially no target product detectable in the control experiment (lane 3), while a weak target product gel band was obtained in self-catalysis experiment (lane 7), thus demonstrating the successful performance of the self-catalysis reaction.
Interestingly, with the concentrations of DNAzyme-2 increased, the differences of target product generations between the control and self-catalysis experiments gradually reduced (Figure 5D). Here, we measured the feedback efficiency using the catalysis ratio, which is the number obtained by comparing the incremental generations of target products in the control and self-catalysis experiments, representing the self-catalysis ability (more details in supporting information section 2, Supplementary Figure S7). From the result of software IMAGE J, the catalysis ratios of varying DNAzyme-2 concentrations (0.05, 0.15, 0.45 and 1.35 μM) were 0.96, 1.18, 0.23, 0.18, respectively. Clearly, the catalysis ratio values have a decreasing tendency with DNAzyme-2 concentration increased. We attribute this phenomenon as, at low DNAzyme-2 concentration, the target products quantities directly induced by initial DNAzyme-2 were rather small, and the quantities caused by feedback trigger DNAzyme-2b were relative high. Therefore, the self-catalysis effect is much easier to be observed at the low DNAzyme-2 concentrations. This speculation was also verified with the self-catalysis simulation model in Supplementary Data Section 3 (Supplementary Figure S11).
Furthermore, the self-catalysis effect was also confirmed by fluorescence assay, in which a series of DNAzyme-2 concentrations were used as 0.1, 0.2, 0.4, 0.6, 0.8 and 1.0 μM (Figure 5E and Supplementary Figure S6). Here, the self-catalysis ability was represented by using the relative percentage (ΔF). Similar to the gel results, the differences between the self-catalysis and control experiments became smaller, when initial DNAzyme-2 concentrations increased (Figure 5E). Overall, these results verified the performance of feedback reaction based on DNAzyme regulation.
CONCLUSION
In summary, we have developed a system of catalytic DNA logic gates regulated by DNAzymes. In the system, the DNAzyme acts as an input to produce the covalent modification (e.g. cleaving backbone bonds). Then, the circuits respond to the conformational changes and facilitate the entropy-driven strand displacement. Meanwhile, a new DNAzyme can be produced during one cycle of such catalytic DNA circuit. Therefore, DNAzyme and DNA catalyst can participate into the reactions alternatively, thus realizing a cascading catalytic circuit. In this study, a series of basic DNA logic gates were established, including YES, OR and AND. In addition, the two-layer cascading DNA circuit was constructed to demonstrate the scalability and hierarchy of the regulation method. At last, the self-catalysis circuit was also performed, in which the feedback and report functions were simultaneously realized via the DNAzyme-based regulation. Moreover, the concentration-dependent differentiation was found between the self-catalysis and control experiments. Overall, the results demonstrated that the DNAzyme regulation strategy is suitable for regulating complex hierarchical catalytic DNA circuits. In particular, this method also shows a promise for constructing complicated nanodevices and nanorobots. We envision that, combined with other recently developed sensing methods, the resulting cacscading DNA circuits will have further potential applications in related fields of biosensing and molecular engineering.
Supplementary Material
ACKNOWLEDGEMENTS
Authors’ contribution: J.Y. and C.Z. conceived the project, designed experiments, analyzed data and wrote the manuscript. R.W. performed experiments. Z.W. and Y.L. constructed the simulation models. Z.L., L.P. and Q.Z. discussed the experiment results.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program of China [2016YFA0501603 to Z.L.]; Joint Found of the Equipment Pre Research Ministry of Education [6141A02033607 to J.Y.]; Central University, Fundamental Research Funds [2016MS46 to J.Y.]; National Natural Science Foundation of China [61370099, 61472333 to J.Y., 61751203 to C.Z., 61320106005 to L.P.]; Beijing Natural Science Foundation [4182027 to C.Z.]; Beijing Municipal Key R&D Project [Z151100003915081 to Z.L.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Seeman N.C. DNA in a material world. Nature. 2003; 421:427–431. [DOI] [PubMed] [Google Scholar]
- 2. Shlyahovsky B., Li Y., Lioubashevski O., Elbaz J., Willner I.. Logic gates and antisense DNA devices operating on a translator nucleic acid scaffold. ACS Nano. 2009; 3:1831–1843. [DOI] [PubMed] [Google Scholar]
- 3. Zhu J., Zhang L., Dong S., Wang E.. Four-way junction-driven DNA strand displacement and its application in building majority logic circuit. ACS Nano. 2013; 7:10211–10217. [DOI] [PubMed] [Google Scholar]
- 4. Zhang C., Yang J., Jiang S., Liu Y., Yan H.. DNAzyme-based logic gate-mediated DNA self-assembly. Nano Lett. 2016; 16:736–741. [DOI] [PubMed] [Google Scholar]
- 5. De Silva A.P., Uchiyama S.. Molecular logic and computing. Nat. Nanotech. 2017; 2:399–410. [DOI] [PubMed] [Google Scholar]
- 6. Voelcker N.H., Guckian K.M., Saghatelian A., Ghadiri M.R.. Sequence-addressable DNA logic. Small. 2008; 4:427–431. [DOI] [PubMed] [Google Scholar]
- 7. Chandrasekaran A. R., Levchenko O., Patel D. S.. Molly MacIsaac and Ken Halvorsen. Addressable configurations of DNA nanostructures for rewritable memory. Nucleic Acids Res. 2017; 45:11459–11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qian L., Winfree E.. Scaling up digital circuit computation with DNA strand displacement cascades. Science. 2011; 332:1196–1201. [DOI] [PubMed] [Google Scholar]
- 9. Qian L., Winfree E., Bruck J.. Neural network computation with DNA strand displacement cascades. Nature. 2011; 475:368–372. [DOI] [PubMed] [Google Scholar]
- 10. Chirieleison S.M., Allen P.B., Simpson Z.B., Ellington A.D.. Pattern transformation with DNA circuits. Nat. Chem. 2013; 5:1000–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gerasimova Y., Kolpashchikov D.. Towards a DNA nanoprocessor: reusable tile-integrated DNA circuits. Angew. Chem. Int. Ed. 2016; 128:10400–10403. [DOI] [PubMed] [Google Scholar]
- 12. Chatterjee G., Dalchau N., Muscat R. A., Phillips A., Seelig G.. A spatially localized architecture for fast and modular DNA computing. Nat. Nanotech. 2017; 12:920–927. [DOI] [PubMed] [Google Scholar]
- 13. Bhadra S., Ellington A. D.. Design and application of cotranscriptional non-enzymatic RNA circuits and signal transducers. Nucleic Acids Res. 2014; 42:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Douglas S.M., Bachelet I., Church G.M.. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012; 335:831–834. [DOI] [PubMed] [Google Scholar]
- 15. Zhu J., Zhang L., Li T., Dong S., Wang E.. Enzyme-free unlabeled DNA logic circuits based on toehold-mediated strand displacement and split G-quadruplex enhanced fluorescence. Adv. Mater. 2013; 25:2440–2444. [DOI] [PubMed] [Google Scholar]
- 16. Elbaz J., Lioubashevski O., Wang F., Remacle F., Levine R.D., Willner I.. DNA computing circuits using libraries of DNAzyme subunits. Nat. Nanotech. 2010; 5:417–422. [DOI] [PubMed] [Google Scholar]
- 17. Sanchita B., Andrew D.E.. Design and application of cotranscriptional non-enzymatic RNA circuits and signal transducers. Nucleic Acids Res. 2014; 42:e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang D.Y., Winfree E.. Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 2009; 131:17303–17314. [DOI] [PubMed] [Google Scholar]
- 19. Chakraborty B., Sha R., Seeman N.C.. A DNA-based nanomechanical device with three robust states. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:17245–17249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xing Y., Yang Z., Liu D.. A responsive hidden toehold to enable controllable DNA strand displacement reactions. Angew. Chem. 2011; 123:12140–12142. [DOI] [PubMed] [Google Scholar]
- 21. Frezza B.M., Cockroft S.L., Ghadiri M.R.. Modular multilevel circuits from immobilized DNA-based logic gates. J. Am. Chem. Soc. 2007; 129:14875–14879. [DOI] [PubMed] [Google Scholar]
- 22. Zhang C., Shen L., Liang C., Dong Y., Yang J., Xu J.. DNA sequential logic gate using two-ring DNA. ACS Appl. Mater. Interfaces. 2016; 8:9370–9376. [DOI] [PubMed] [Google Scholar]
- 23. Shi L., Peng P., Du Y., Li T.. Programmable i-motif DNA folding topology for a pH-switched reversible molecular sensing device. Nucleic Acids Res. 2017; 45:4306–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang F., Orbach R., Willner I.. Detection of metal ions (Cu2+, Hg2+) and cocaine by using ligation DNAzyme machinery. Chem. Eur. J. 2012; 18:16030–16036. [DOI] [PubMed] [Google Scholar]
- 25. Georg S., David S., David Y.Z., Erik W.. Enzyme-free nucleic acid logic circuits. Science. 2006; 314:1585–1588. [DOI] [PubMed] [Google Scholar]
- 26. David Y.Z., Andrew J.T., Bernard Y., Erik W.. Engineering entropy-driven reactions and networks catalyzed by DNA. Science. 2007; 318:1121–1125. [DOI] [PubMed] [Google Scholar]
- 27. Xia F., Zuo X., Yang R., White R.J., Xiao Y., Kang D., Gong X., Lubin A.A., Vallee-BelÀisle A., Yuen J.D.. Label-free, dual-analyte electrochemical biosensors: a new class of molecular-electronic logic gates. J. Am. Chem. Soc. 2010; 132:8557–8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu J., Zhang L., Li T., Dong S., Wang E.. Enzyme-free unlabeled DNA logic circuits based on toehold-mediated strand displacement and split G-quadruplex enhanced fluorescence. Adv. Mater. 2013; 25:2440–2444. [DOI] [PubMed] [Google Scholar]
- 29. Chen X. Expanding the rule set of DNA circuitry with associative toehold activation. J. Am. Chem. Soc. 2012; 134:263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang F., You M., Han D., Xiong X., Liang H., Tan L.. DNA branch migration reactions through photocontrollable toehold formation. J. Am. Chem. Soc. 2013; 135:7967–7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Elbaz J., Wang F., Remacle F., Willner I.. pH-Programmable DNA logic arrays powered by modular DNAzyme libraries. Nano Lett. 2012; 12:6049–6054. [DOI] [PubMed] [Google Scholar]
- 32. Pan L., Wang Z., Li Y., Xu F., Zhang Q., Zhang C.. Nicking enzyme-controlled toehold regulation for DNA logic circuits. Nanoscale. 2017; 9:18223–18228. [DOI] [PubMed] [Google Scholar]
- 33. Lu L., Zhang X., Kong R., Yang B., Tan W.. A ligation-triggered DNAzyme cascade for amplified fluorescence detection of biological small molecules with zero-background signal. J. Am. Chem. Soc. 2011; 133:11686–11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang S., Yue L., Shpilt Z., Cecconello A., Kahn J.S., Lehn J., Willner I.. Controlling the catalytic functions of DNAzymes within constitutional dynamic networks of DNA nanostructures. J. Am. Chem. Soc. 2017; 139:9662–9671. [DOI] [PubMed] [Google Scholar]
- 35. Endo M., Takeuchi Y., Suzuki Y., Emura T., Hidaka K., Wang F., Willner I., Sugiyama H.. Single-molecule visualization of the activity of a Zn2+-dependent DNAzyme. Angew. Chem. Int. Ed. 2015; 54:10550–10554. [DOI] [PubMed] [Google Scholar]
- 36. Zhang Z., Balogh D., Wang F., Willner I.. Smart mesoporous SiO2 nanoparticles for the DNAzyme-induced multiplexed release of substrates. J. Am. Chem. Soc. 2013; 135:1934–1940. [DOI] [PubMed] [Google Scholar]
- 37. Wang F., Elbaz J., Teller C., Willner I.. Amplified detection of DNA through an autocatalytic and catabolic DNAzyme-mediated process. Angew. Chem. Int. Ed. 2011; 50:295–299. [DOI] [PubMed] [Google Scholar]
- 38. Po-Jung J.H., Jenny L., Jing C., Mahsa V., Liu J.. Ultrasensitive DNAzyme beacon for lanthanides and metal speciation. Anal. Chem. 2014; 86:1816–1821. [DOI] [PubMed] [Google Scholar]
- 39. Chen Y., Robert M.C.. DNAzyme footprinting: detecting protein-aptamer complexation on surfaces by blocking DNAzyme cleavage activity. J. Am. Chem. Soc. 2013; 135:2072–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang F., Elbaz J., Teller C., Willner I.. Amplified detection of DNA through an autocatalytic and catabolic DNAzyme-mediated process. Angew. Chem. Int. Ed. 2011; 50:295–299. [DOI] [PubMed] [Google Scholar]
- 41. Zhang J., Wang L., Zhang H., Boey F., Song S., Fan C.. Aptamer-based multicolor fluorescent gold nanoprobes for multiplex detection in homogeneous solution. Small. 2010; 6:201–204. [DOI] [PubMed] [Google Scholar]
- 42. Kong R., Zhang X., Chen Z., Meng H., Song Z., Tan W., Shen G., Yu R.. Unimolecular catalytic DNA biosensor for amplified detection of l-histidine via an enzymatic recycling cleavage strategy. Anal. Chem. 2011; 83:7603–7607. [DOI] [PubMed] [Google Scholar]
- 43. Shlyahovsky B., Li Y., Lioubashevski O., Elbaz J., Willner I.. Logic gates and antisense DNA devices operating on a translator nucleic acid scaffold. ACS Nano. 2009; 3:1831–1843. [DOI] [PubMed] [Google Scholar]
- 44. Bi S., Yan Y., Hao S., Zhang S.. Colorimetric logic gates based on supramolecular DNAzyme structures. Angew. Chem. Int. Ed. 2010; 49:4438–4442. [DOI] [PubMed] [Google Scholar]
- 45. Moshe M., Elbaz J., Willner I.. Sensing of UO22+ and design of logic gates by the spplication of supramolecular constructs of ion-dependent DNAzymes. Nano Lett. 2009; 9:1196–1200. [DOI] [PubMed] [Google Scholar]
- 46. Elbaz J., Wang F., Remacle F., Willner I.. pH-programmable DNA logic arrays powered by modular DNAzyme libraries. Nano Lett. 2012; 12:6049–6054. [DOI] [PubMed] [Google Scholar]
- 47. Brown C.W. III, Lakin M.R., Horwitz E.K., Fanning M.L., West H.E., Stefanovic D., Graves S.W.. Signal propagation in multi-layer DNAzyme cascades using structured chimeric substrates. Angew. Chem. Int. Ed. Engl. 2014; 53:7183–7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brown C.W. III, Lakin M.R., Stefanovic D., Graves S.W.. Catalytic molecular logic devices by DNAzyme displacement. Chembiochem. 2014; 15:950–954. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.