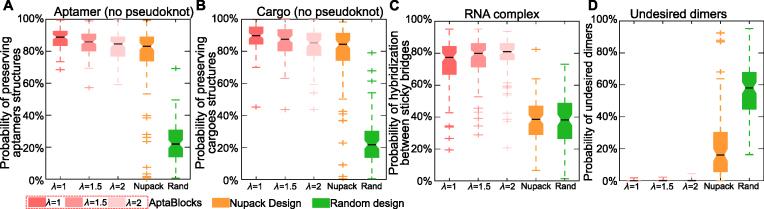
Figure 2.
Comparison of the competing algorithms. (A) Comparison of conserving the secondary structure of aptamers. The probability of sticky bridges being unpaired is computed using a secondary structure model without pseudoknots. (B) Comparison of conserving the secondary structure of cargoes. The probability of sticky bridges being unpaired is computed using a secondary structure model without pseudoknots. (C) Comparison of binding affinity. The binding affinity is approximated by computing the probability of hybridization between sticky bridges using an RNA-RNA interaction model. (D) Comparison on incurring undesired dimers. The probability of undesired dimers is computed by our proposed model in Supplementary Materials B. Larger λ indicates that AptaBlocks focuses more on increasing the melting temperature of sticky bridges.