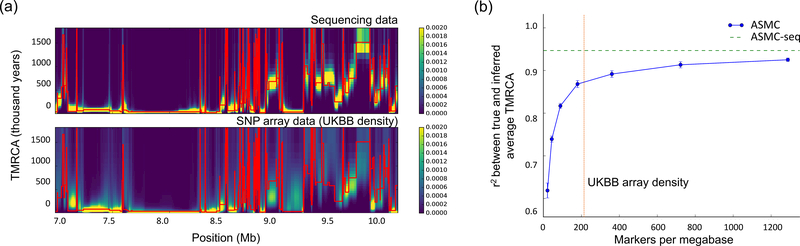
Figure 1. ASMC accuracy in coalescent simulations.
(a) Sample posterior decoding of TMRCA along a 3 Mb segment for a pair of simulated individuals with ASMC run on WGS data (top) and on SNP array data (bottom). Red lines represent the true TMRCA, while the heat map represents the inferred posterior distribution. Posterior density tends to concentrate more tightly around the true TMRCA when WGS data are analyzed, due to the higher density of polymorphic variants. Posterior estimates using SNP array data are more dispersed for distant TMRCA, but remain highly concentrated for recent TMRCA. (b) Accuracy (r2 between true and inferred average TMRCA) as a function of marker density. TMRCA are inferred using the posterior mean obtained by ASMC at each site. ASMC-seq represents the accuracy obtained using ASMC on WGS data. The red vertical line indicates marker density in the UK Biobank data set. Errors bars represent standard errors. Dots and error bars represent the average and its SE from 10 independent simulations. Numerical results are reported in Supplementary Table 11.