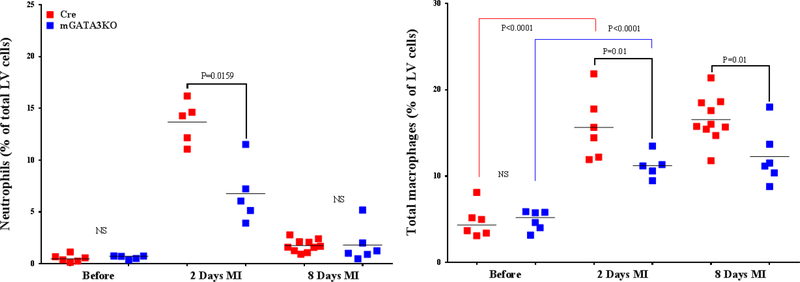
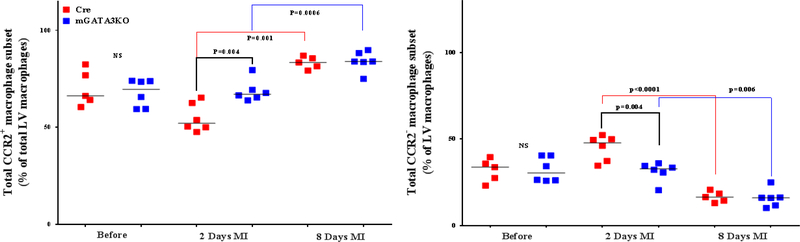
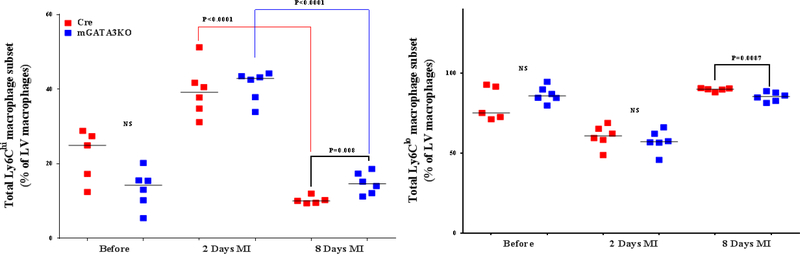
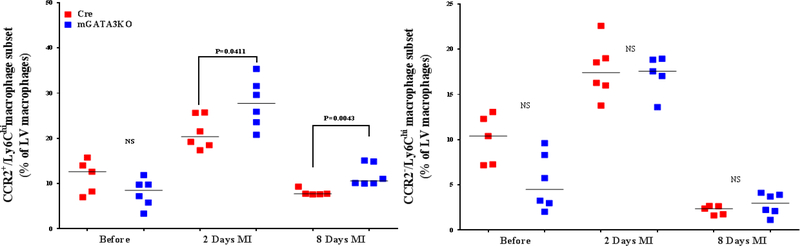
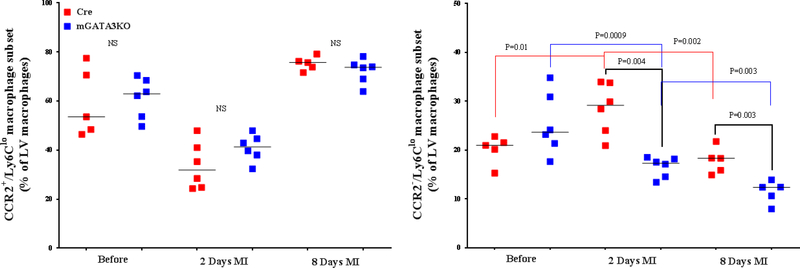
Figure 3. Phenotype of Monocytes/Macrophages in the LV of the 2 Mouse Genotypes.
LVs from the 2 mouse genotypes before and at the indicated days after MI were digested, stained with appropriate antibodies, and analyzed by flow cytometry to determine monocyte and macrophage phenotypes. Each panel depicts a specific cell subset based on the expression of cell markers. (A) neutrophils (left) and total macrophages (right); (B) total CCR2+ (left) and CCR2− macrophage subsets (right); (C) total Ly6Chi (left) and total Ly6Clo macrophage subsets (right); (D) CCR2+/Ly6Chi (left) and CCR2−/Ly6Chi macrophage subsets (right); (E) CCR2+/Ly6Clo (left) and CCR2−/Ly6Clo macrophage subsets (right). Each data point represents 1 mouse/genotype. Total CCR2+, CCR2−, Ly6Chi, or Ly6Clo cells were calculated by addition of the frequencies of each cell subset. Data from the 2 groups (Cre versus mGATA3KO) before and after MI were analyzed by Student t-test. Each data point represents 1 mouse; therefore, the number of animals in each group is equal to the number of dots/group. Abbreviations as in Figure 1.