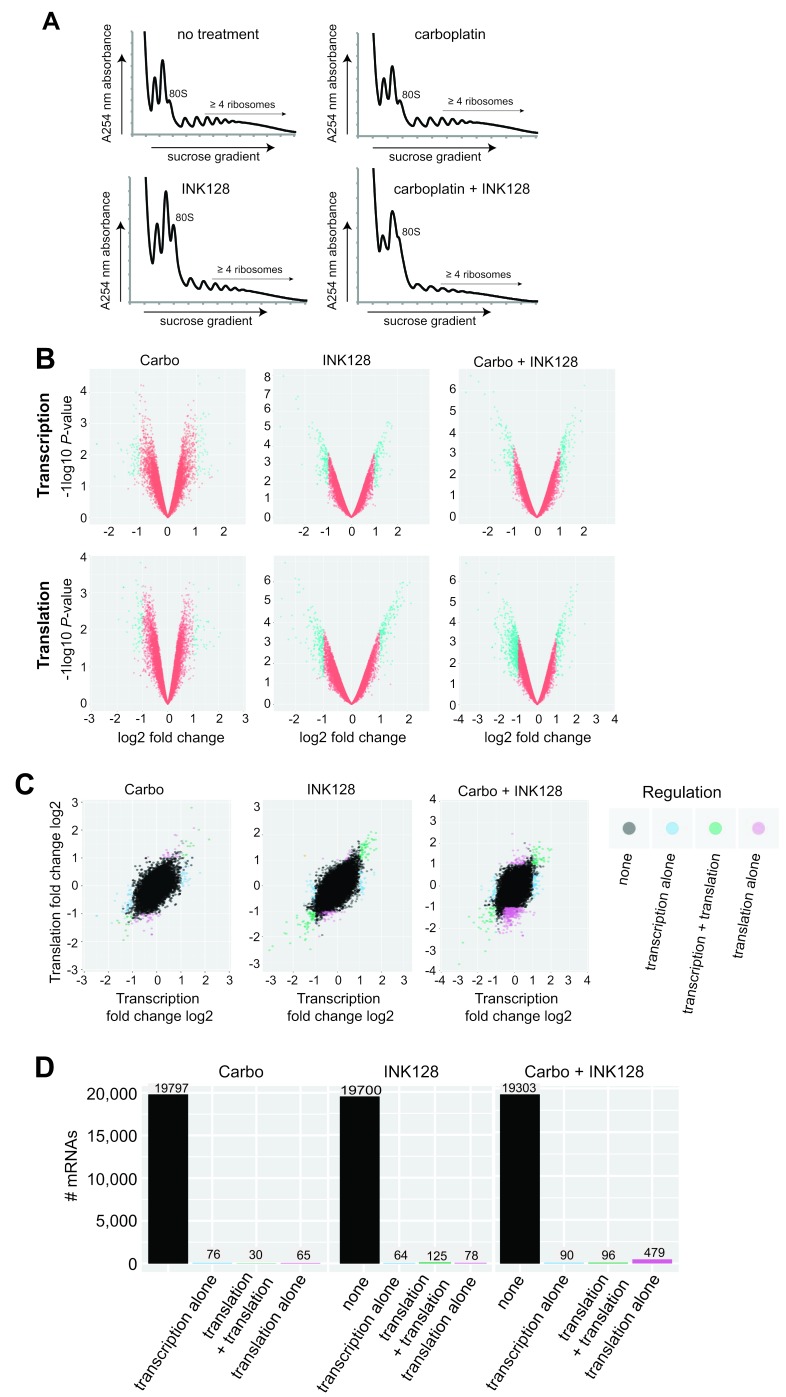
Figure 3. Transcriptomic and translatomic analysis of carboplatin re-sensitization by mTORC1/2 inhibition.
A. Absorbance profiles of ribosome subunits and polysomes for untreated and all treatment groups. OVCAR-3 cells were treated with DMSO as control, 1 µM carboplatin, 0.25 µM INK128, or both, as described in Material and Methods. Cycloheximide (100 µg/ml) was added, polysomes prepared and sorted by centrifugation through 10-50% sucrose gradients using equal RNA amounts and equal volume fractions collected while simultaneously monitoring absorbance at 254 nm, as described [38]. B. Volcano plots demonstrating transcriptional and translational alterations plotted as log2 fold changes against log10 P-values for all three treatment conditions. C. Log2 scatter plots of transcriptomic and translatomic results for all treatment groups (carboplatin alone, INK128 alone, combination treatment) analyzed for altered transcription, transcription + translation, or translation alone (translation efficiency). Two complete sets of independently performed studies were used to develop transcriptome and translatome data sets for analysis. D. Histogram representation of number of mRNAs out of total mRNAs altered in all three treatment conditions for transcription, transcription + translation, or translation alone (translation efficiency).