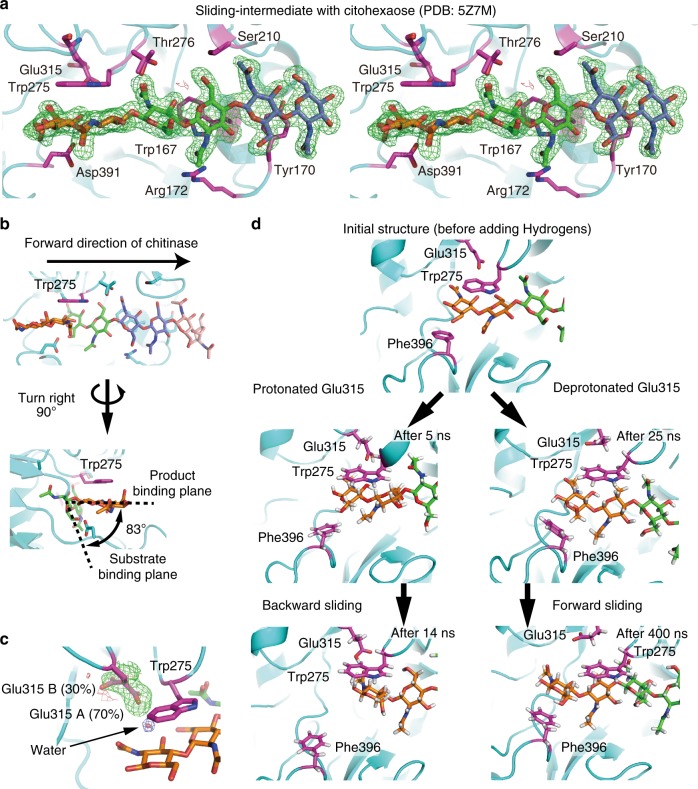
Fig. 4.
Structural and MD simulation analyses of chain sliding of SmChiA. a Cross-eyed stereo view of the omit map of chitohexaose molecule in the Sliding-intermediate structure (PDB ID: 5Z7M) shown at 3σ; green: positive; red: negative. Substrate-interacting amino acid residues are shown by purple. b Product- and substrate-binding planes in the Michaelis complex-like structure of inactive D313A mutant (PDB ID: 1EIB); inter plane angle is 83°. c Omit map of Glu315 side chain in the Sliding-intermediate structure shown at 3σ; green: positive; red: negative. 2Fobs − Fcalc map of water molecule is also shown in 1.5σ (arrow). d Snap shots of MD simulation showing forward and backward chain slidings from the Sliding-intermediate structure. Structures before (25 ns) and after (400 ns) forward sliding with deprotonated Glu315 are shown on the right, and those before (5 ns) and after (14 ns) backward sliding with protonated Glu315 are shown on the left. Trp275, Glu315, and Phe396 were shown by stick. Structures were aligned with Trp275 as the centre of view