Fig. 4.
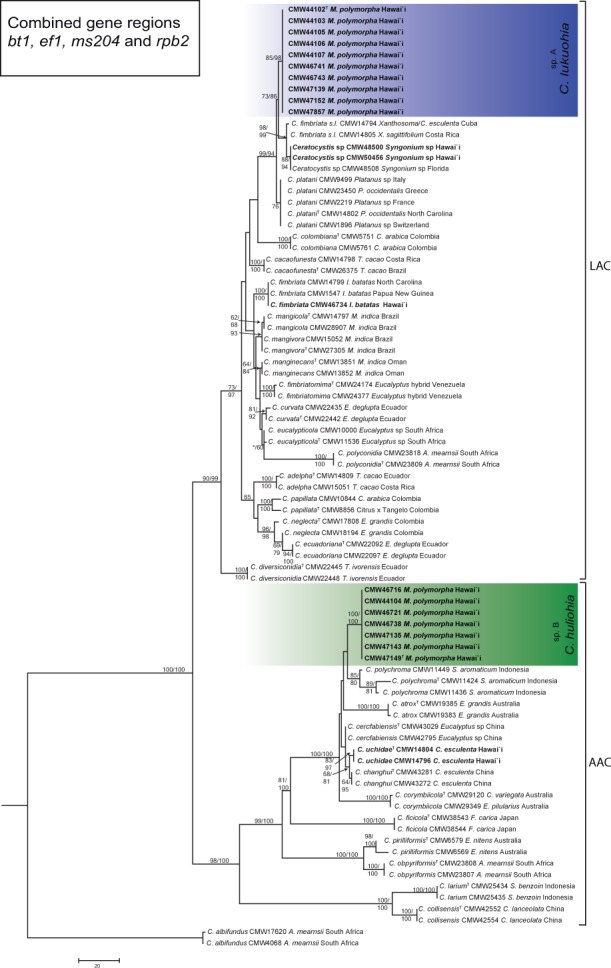
Maximum parsimony tree based on the combined dataset of four gene regions (bt1, tef1, ms204 and rpb2) for species in the Latin American clade (LAC) and the Asian-Australian clade (AAC) of Ceratocystis. Isolates sequenced in this study from M. polymorpha in Hawai’i are highlighted in blue in the LAC, as C. lukuohia (sp. A), and in green in the AAC, as C. huliohia (sp. B). Bootstrap values > 60 % for MP/ML are presented at the branches. Bootstrap values lower than 70 % are indicated with *. Ceratocystis albifundus (CMW 4068) from the African clade was used as the outgroup.
