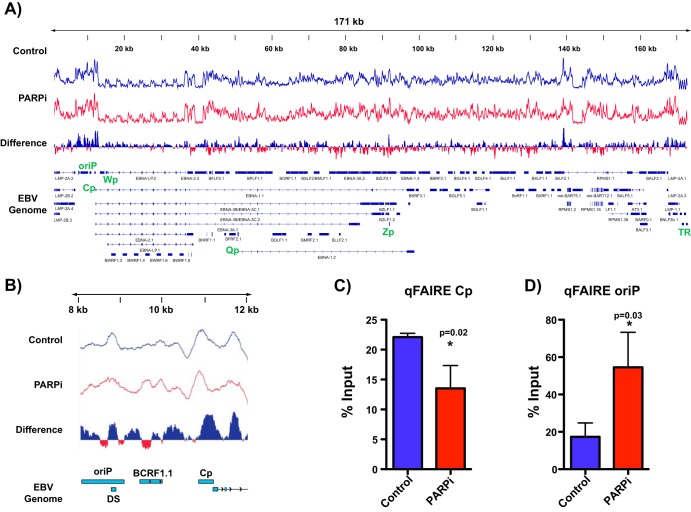
FIG 4.
PARP inhibition results in more tightly packed chromatin at the Cp promoter. (A) Formaldehyde-assisted isolation of regulatory element sequencing (FAIRE-seq) to assess chromatin accessibility across the EBV genome in LCLs before (control; blue track) and after inhibition of PARP activity (PARPi; red track). By differential analysis, blue peaks represent regions in which open chromatin is enriched in the control, and red peaks represent regions with more open chromatin in the PARPi sample. (B) Zoomed image of FAIRE-seq at the latent Cp locus in LCLs, demonstrating the packing of chromatin at Cp after PARP inhibition. (C) Independent FAIRE-qPCR validation at Cp and OriP in untreated or olaparib-treated (PARPi) LCLs. qPCR data are presented as percentages of input DNA. Results are representative of three independent experiments and show means ± standard deviations. Statistical significance, as determined by Student's t test, is indicated by asterisks and P value.