Epstein-Barr virus (EBV) is the first identified human tumor virus and is associated with a range of human cancers. During EBV-induced lymphomas, the essential viral latent proteins modify the expression of cell cycle-related proteins to disturb the cell cycle process, thereby facilitating the proliferative process. The essential EBV nuclear antigen 3C (EBNA3C) plays an important role in EBV-mediated B-cell transformation. Here we show that EBNA3C stabilizes cyclin D2 to regulate cell cycle progression. More specifically, EBNA3C directly binds to cyclin D2, and they colocalize together in nuclear compartments. EBNA3C enhances cyclin D2 stability by inhibiting its ubiquitin-dependent degradation and significantly promotes cell proliferation in the presence of cyclin D2. Our results provide novel insights into the function of EBNA3C on cell progression by regulating the cyclin D2 protein and raise the possibility of the development of new anticancer therapies against EBV-associated cancers.
KEYWORDS: cyclin D2, EBNA3C, Epstein-Barr virus, cell proliferation
ABSTRACT
Cell cycle regulation is one of the hallmarks of virus-mediated oncogenesis. Epstein-Barr virus (EBV)-induced lymphomas express a repertoire of essential viral latent proteins that regulate expression of cell cycle-related proteins to dysregulate this process, thereby facilitating the proliferation of infected cells. We now demonstrate that the essential EBV latent protein 3C (EBNA3C) stabilizes cyclin D2 to regulate cell cycle progression. More specifically, EBNA3C directly binds to cyclin D2 and they colocalize together in nuclear compartments. We show that EBNA3C regulates the promoter of cyclin D2 through cooperation with master transcription factor Bcl6 and enhances its stability by inhibiting its ubiquitin-dependent degradation. EBNA3C also promoted cell proliferation in the presence of cyclin D2, suggesting that cyclin D2 contributes to EBNA3C-mediated cell cycle progression. These results provide new clues as to the role of this essential viral latent protein and its ability to regulate expression of cellular factors, which drives the oncogenic process.
IMPORTANCE Epstein-Barr virus (EBV) is the first identified human tumor virus and is associated with a range of human cancers. During EBV-induced lymphomas, the essential viral latent proteins modify the expression of cell cycle-related proteins to disturb the cell cycle process, thereby facilitating the proliferative process. The essential EBV nuclear antigen 3C (EBNA3C) plays an important role in EBV-mediated B-cell transformation. Here we show that EBNA3C stabilizes cyclin D2 to regulate cell cycle progression. More specifically, EBNA3C directly binds to cyclin D2, and they colocalize together in nuclear compartments. EBNA3C enhances cyclin D2 stability by inhibiting its ubiquitin-dependent degradation and significantly promotes cell proliferation in the presence of cyclin D2. Our results provide novel insights into the function of EBNA3C on cell progression by regulating the cyclin D2 protein and raise the possibility of the development of new anticancer therapies against EBV-associated cancers.
INTRODUCTION
As the first identified human tumor virus, Epstein-Barr virus (EBV) has been studied for more than 50 years and is associated with a range of different cancers (1–3). It was discovered from African Burkitt's lymphoma tissue, is thought to be the most common persistent and asymptomatic virus, and infects greater than 90% of the world's population (4, 5). Infection of B cells by EBV leads to aberrant cell proliferation and development of immortalized lymphoblastoid cell lines under specific in vitro conditions. During this type of infection, referred to as latency III, EBV latent infection is established, and its associated latent genes, including the genes for six latent EBV nuclear antigens (EBNAs; EBNA1, EBNA2, EBNA3A, EBNA3B, EBNA3C, EBNA-LP) and three latent membrane proteins (LMPs; LMP-1, LMP-2A, LMP-2B), as well as EBV-encoded RNAs (EBERs) and the BARTs (6), are expressed. Furthermore, molecular genetic analyses have found that EBNA2, EBNA3A, EBNA3C, EBNA-LP, and LMP-1 are essential for EBV-induced in vitro immortalization of human primary B cells (7–11).
Different from normal tissues, cancer cells lose control of the cell cycle or cell growth, which leads to unlimited cell proliferation (12). As critical components of cell cycle progression, cyclin D family members are typically dysregulated in cancers, which makes them valuable therapeutic targets for cancer therapy (13). Cyclin D proteins bind and activate cyclin-dependent kinase 4 (CDK4) or CDK6 to regulate downstream targets, especially the famous tumor suppressor retinoblastoma protein (Rb), and further activate or inhibit E2F transcription factors (14–16). Therefore, the classical cyclin/cyclin-dependent kinase-Rb-E2F pathway demonstrates the critical functions of cyclin D proteins in the carcinogenic process. Notably, the overexpression of cyclin D1 has been found in breast and many other cancers (13). The stabilization of cyclin D2 has also been shown to be a major contributor to phosphatidylinositol 3-kinase–AKT-related megalencephaly syndrome (17). Further, overexpression of cyclin D3 is related to a number of lymphoid-associated malignancies (13). Compared to the intense amount of studies on cyclin D1, very few studies have been completed on cyclin D2 or D3 (18).
The oncoproteins encoded by EBV have been shown to control the cell cycle machinery through regulation of many cellular signaling pathways during EBV infection. For example, the EBV Zta transactivator induces cell cycle arrest in G0-G1 by targeting p53, p21, p27, and pRb in epithelial cells (19). Both LMP-1 and LMP-2A can downregulate the expression of the forkhead transcription factor FoxO1, which ultimately increases cyclin D2 expression (20). MicroRNAs encoded by the EBV BHRF1 locus can also regulate cell cycle progression (21), and the upregulation of cyclin D2 in Mutu I EBV-positive cells suggested that EBV may influence its expression (22). In addition, the EBNA3 family members expressed during latent infection can also facilitate B-cell transformation by controlling critical nodes in the regulatory network of host gene transcription. EBNA3C is one of the essential latent antigens that interacts with numerous host transcriptional factors, further regulating the virus-host interaction network (23). Our previous studies have identified many cellular factors that associate with EBNA3C, including Nm23-H1 (24), Rb (25), p53 (26), E2F1 (27), E2F6 (28), and Bcl6 (29). Previously, one study indicated that EBNA3C inhibits p16INK4A-mediated Rb dephosphorylation to facilitate cell cycle progression (30), and other reports showed that EBNA3C can stimulate cyclin A-dependent kinase activity (31, 32).
Our previous study showed that EBNA3C can stabilize and enhance cyclin D1 activity, thereby promoting G1/S transition in EBV-transformed cells (33). However, whether the essential latent EBV nuclear antigen EBNA3C has any roles in regulating cyclin D2 activity during the cell cycle is largely unknown. Cyclin D2 is also highly expressed in late G1 phase and facilitates the G1/S transition, especially in EBV-transformed lymphoblastoid cells (33). This suggests a crucial function of EBV latent nuclear proteins on cell cycle progression by promoting cyclin D2 expression. Here, we further investigated the effects of EBNA3C on cyclin D2 expression in EBV-infected B cells. These results demonstrate that EBNA3C is required to stabilize cyclin D2 and that it contributes to the oncogenic process through regulating cyclin D2-mediated cell progression.
RESULTS
EBNA3C expression leads to increased levels of cyclin D2.
Cyclin D proteins are homologous within their conserved domains, but their functions are not identical across the three proteins (13, 34–36). To further investigate the differences in functions of these different cyclin D proteins, Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood City, CA) was used to analyze their regulatory networks. Modification of individual cyclin D protein expression in silico provided predictions of the expression of the other two cyclin D proteins. For example, cyclin D2 expression was downregulated when the cyclin D1 level was upregulated, whereas cyclin D3 expression showed no obvious change (data not shown). Interestingly, cyclin D2 expression showed a trend opposite that for cyclin D1 or D3 protein expression in silico (data not shown). These in silico analyses demonstrate that cyclin D2 likely executes specific functions that are different from those of cyclin D1 and cyclin D3.
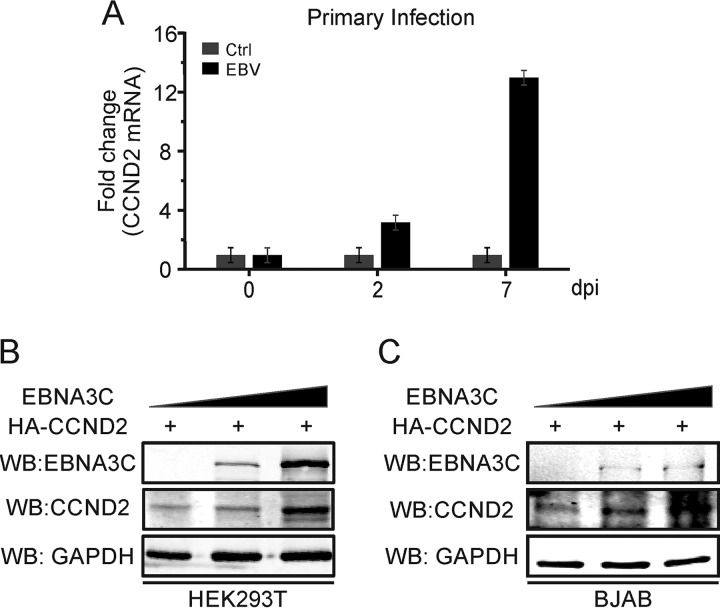
Considering the potential specificity of cyclin D2 compared to the other cyclin D proteins, we wanted to further determine its role during EBV infection. Peripheral blood mononuclear cells (PBMCs) were infected with wild-type EBV and collected at the time points indicated below to monitor cyclin D2 mRNA expression levels. The results demonstrated that cyclin D2 mRNA expression was significantly upregulated after EBV infection of PBMCs compared with that in the mock EBV infection control (Fig. 1A). Importantly, cyclin D2 levels began to increase by 48 h postinfection and increased to more than 12-fold over those in the mock-infected control by 7 days (Fig. 1A). This suggests that cyclin D2 likely plays a role in EBV primary infection and that its expression is likely regulated by virally encoded proteins.
FIG 1.
EBNA3C expression leads to increased levels of cyclin D2 (CCND2). (A) Ten million PBMCs were infected with wide-type EBV or mock infected. Then, the infected cells were collected at the indicated time points, and cyclin D2 mRNAs were detected using real-time PCR. GAPDH was used as the internal control. dpi, day postinfection. (B, C) Ten million HEK293T (B) or BJAB (C) cells were cotransfected with an increased dose of Myc-tagged EBNA3C and Flag-tagged cyclin D2. At 48 h posttransfection, the EBNA3C and cyclin D2 proteins were detected by Western blotting. GAPDH was used as a control protein.
Among the EBV latent proteins, EBNA1 was not considered the main latent antigen responsible for cyclin D2 expression, because cyclin D2 expression was dramatically enhanced in Burkitt's lymphoma Mutu III cells compared to that in Mutu I cells, which are EBV positive (33). In addition, our previous results also indicated that EBNA3C can stabilize the cyclin D1 protein (33). Therefore, it was reasonable to predict that EBNA3C may also regulate cyclin D2 expression because of the tight conservation of the cyclin D family members (13). To further investigate the potential effects of EBNA3C on the regulation of the cyclin D2 protein, increasing amounts of EBNA3C plasmids were expressed in HEK293T or BJAB cells with cyclin D2 using heterologous systems. After 48 h, Western blot analysis showed that cyclin D2 stability was enhanced in the presence of EBNA3C and that the effect was dose responsive (Fig. 1B and C). These results clearly indicate that EBNA3C is a major contributor to the increased stability of the cyclin D2 protein.
EBNA3C forms a complex with cyclin D2 in cells.
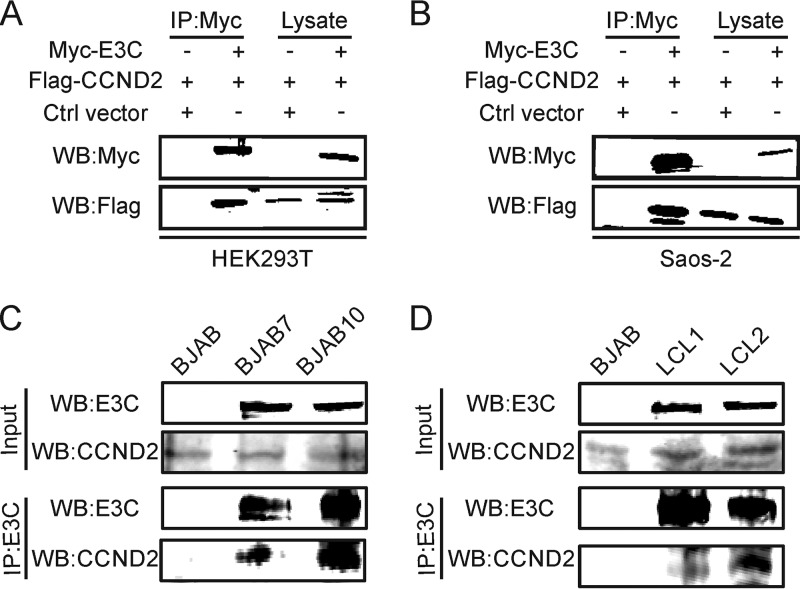
To determine if EBNA3C interacts with cyclin D2, Myc-tagged EBNA3C was transfected into Saos-2 or HEK293T cells with or without Flag-tagged cyclin D2. The transfected cells were harvested at 48 h posttransfection. Coimmunoprecipitation (co-IP) assays were performed using anti-Myc antibody from the 9E10 hybridoma. Immune complexes were fractionated and Western blotted for detection of Myc-tagged EBNA3C and Flag-tagged cyclin D2. The results showed that EBNA3C can associate with cyclin D2 in immune complexes in both epithelial cell and B-cell backgrounds (Fig. 2A and B). To further corroborate their interaction in B cells, BJAB cells and BJAB7 and BJAB10 cells stably expressing EBNA3C were used to further examine their association in these cellular complexes. The results of the coimmunoprecipitation experiments showed that EBNA3C is definitely associated with cyclin D2 in these B-cell lines stably expressing EBNA3C (Fig. 2C). Similarly, the results of coimmunoprecipitation experiments performed using two physiologically relevant EBV-transformed cell lines, lymphoblastoid cell line 1 (LCL1) and LCL2, further support an association of EBNA3C and cyclin D2 in molecular complexes in an EBV-transformed physiologically relevant cell background (Fig. 2D).
FIG 2.
EBNA3C forms a complex with cyclin D2 in cells. (A, B) About 10 million HEK293T (A) or Saos-2 (B) cells were cotransfected with Myc-tagged EBNA3C and Flag-tagged CCND2. Then, cell lysates from the transfected cells were immunoprecipitated with Myc-specific antibody and detected using Western blotting. (C, D) Cell lysates from the indicated B cells (BJAB, BJAB7, BJAB10, LCL1, and LCL2 cells) were immunoprecipitated with EBNA3C-specific antibody and analyzed by Western blotting.
EBNA3C and cyclin D2 colocalize in nuclear compartments.
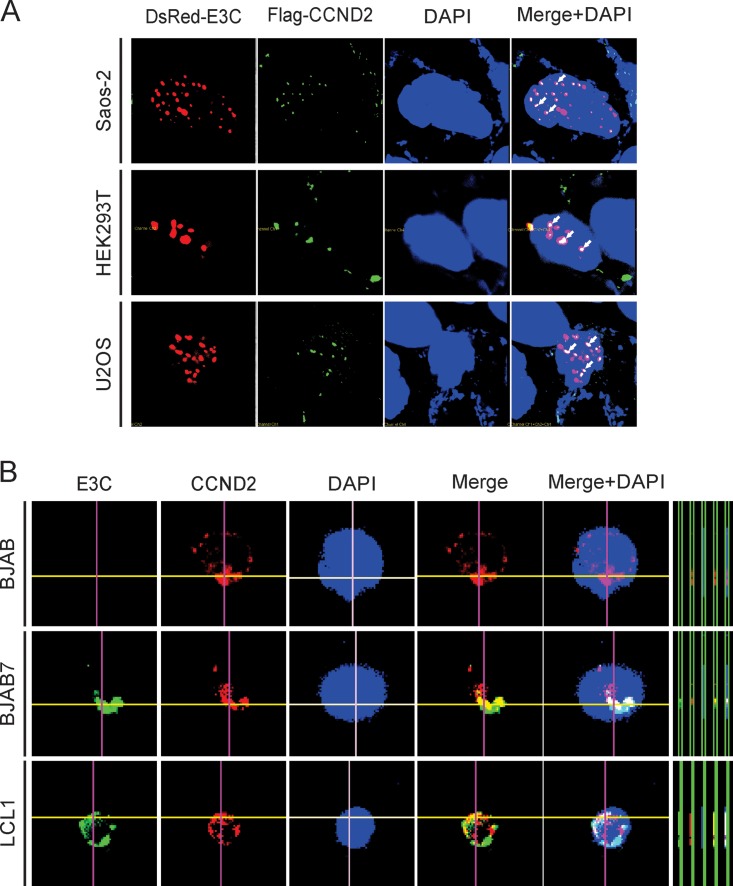
To investigate the localization of EBNA3C and cyclin D2 in human cells, immunofluorescence assays were performed to identify the cellular compartments in which they localize. Previously, studies have shown that EBNA3C, as well as cyclin D2, is located in nuclear compartments (28, 37, 38). However, no information exists about their association in these compartments. Our results now show that EBNA3C and cyclin D2 can colocalize in nuclear compartments of human cells expressing EBNA3C and cyclin D2 from a heterologous promoter (Fig. 3A). In Saos-2, HEK293T, and U2OS cells, both EBNA3C and cyclin D2 showed positive punctate nuclear signals with a high degree of overlap. This suggests that they colocalized in similar nuclear compartments (Fig. 3A). To further determine whether these results were also the case in B cells, we used BJAB cells, BJAB7 cells stably expressing EBNA3C, and LCL1, an EBV-transformed B-cell line, to investigate their colocalization. Similarly, the signals for EBNA3C and cyclin D2 also colocalized in nuclear compartments in B cells (Fig. 3B). These studies indicate that EBNA3C colocalizes with cyclin D2 and may regulate its functions in EBV-transformed lymphoblastoid cells.
FIG 3.
EBNA3C and cyclin D2 colocalize in nuclear compartments. (A) HEK293T, Saos-2, and U2OS cells were cotransfected with Flag-tagged cyclin D2 and DsRed-tagged EBNA3C. The transfected cells were collected after 48 h of transfection, fixed, and incubated with mouse anti-Flag (M2) antibody and Alex Fluor 488-conjugated secondary antibody. DsRed-tagged EBNA3C was directly detected by fluorescence microscope. (B) BJAB, BJAB7, and LCL1 cells were semi-air dried and kept on slides. Endogenous EBNA3C (E3C) or cyclin D2 was incubated with its specific antibody, followed by the corresponding secondary antibodies. Nuclei were stained with DAPI. The images were captured with an Olympus FluoView FV300 microscope and analyzed using its FluoView software.
EBNA3C inhibits cyclin D2 ubiquitination, enhancing its stability.
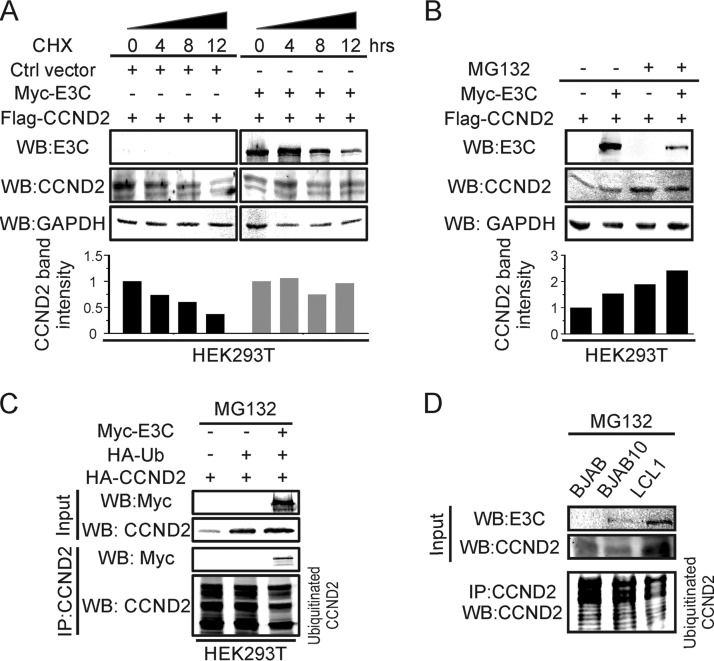
The above-described results show that EBNA3C expression contributes to increased levels of cyclin D2 during primary infection. However, we wanted to further investigate whether this effect is related to the levels of cyclin D2 in infected cells. First, we monitored the stability of cyclin D2 in the absence or presence of EBNA3C after treatment with cycloheximide (CHX) at different time points (Fig. 4A). The results showed that cyclin D2 is more stable in the presence of EBNA3C than the EBNA3C-negative control after inhibition of translation by incubation with CHX at 0, 4, 8, and 12 h posttreatment. Therefore, we wanted to further determine the stability of cyclin D2 by investigating its association with the ubiquitin degradation pathway. We then cotransfected EBNA3C and cyclin D2 and incubated them with or without MG132. The results showed that EBNA3C stabilizes cyclin D2 even without MG132 treatment (Fig. 4B). Importantly, with MG132 treatment, EBNA3C further promoted the stability of cyclin D2 (Fig. 4B). This suggests that EBNA3C may enhance cyclin D2 stability by inhibiting its ubiquitination. To verify this hypothesis, EBNA3C and cyclin D2 expression constructs were transfected together with ubiquitin into cells. At 24 h posttransfection, the cells were incubated with MG132 for another 12 h. As shown, the results indicated that the ubiquitination of cyclin D2 was reduced in the presence of EBNA3C (Fig. 4C). In addition, EBNA3C also inhibited cyclin D2 ubiquitination in MG132-treated B cells (Fig. 4D), suggesting that EBNA3C promoted cyclin D2 stability by suppressing its degradation through the ubiquitin degradation system.
FIG 4.
EBNA3C inhibits cyclin D2 ubiquitination, enhancing its stability. (A) HEK293T cells were cotransfected with Myc-tagged EBNA3C and Flag-tagged cyclin D2. At 36 h posttransfection, the transfected cells were incubated with cycloheximide (CHX) for the indicated times and then lysed, and Western blot analysis was performed. (B) HEK293T cells were cotransfected with Flag-tagged cyclin D2 with or without Myc-tagged EBNA3C. Then, these cells were treated with MG132 or not treated, as shown. Thirty-six hours after transfection, the expression of the targeted protein was analyzed by Western blotting. (C) The indicated plasmids were transfected into HEK293T cells. The transfected cells were incubated with MG132 for 16 h and harvested for Western blot analysis. (D) BJAB, BJAB10, and LCL1 cells were incubated with dimethyl sulfoxide or MG132 for 16 h and collected to detect EBNA3C and cyclin D2 expression. GAPDH was used as a protein control.
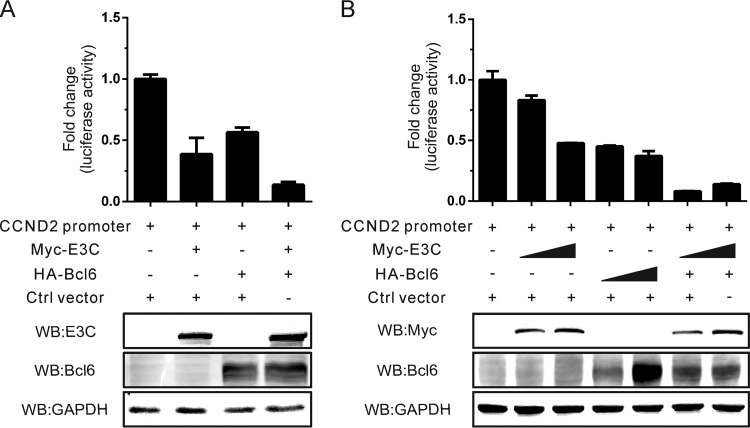
EBNA3C can cooperate with Bcl6 to repress the cyclin D2 promoter.
To further examine whether EBNA3C may regulate the mRNA expression of cyclin D2, dual-luciferase reporter assays were performed using the cyclin D2 promoter luciferase construct. Further, Bcl6 was previously shown to function as a transcription regulator which modulates the activity of the cyclin D2 promoter (39, 40). Therefore, we investigated whether they can cooperate in the regulation of cyclin D2 gene transcription. However, the dual-luciferase reporter assays using the cyclin D2 promoter luciferase construct demonstrated that EBNA3C, similarly to Bcl6, also suppressed the cyclin D2 promoter activity (Fig. 5A). Interestingly, the inhibition was enhanced when they were expressed together, suggesting cooperation (Fig. 5A). However, this effect was not as impressive with increasing amounts of EBNA3C (Fig. 5B). The fact that EBNA3C further enhanced the inhibition of Bcl6-mediated cyclin D2 promoter activity suggested that EBNA3C may also associate with Bcl6 to bind competitively to the promoter or may directly affect Bcl6 expression.
FIG 5.
EBNA3C can cooperate with Bcl6 to repress the cyclin D2 promoter. (A) HEK293T cells were cotransfected with the cyclin D2 promoter, Myc-tagged EBNA3C, HA-tagged Bcl6, the control vector, as well as the Renilla luciferase-expressing plasmid (pRL-TK) and then lysed after 36 h of transfection, and a dual-luciferase reporter assay was performed. (B) HEK293T cells were transfected with the indicated plasmids, and the luciferase reporter assays were performed as mentioned in the text. The expression of transfected plasmids was monitored by Western blotting.
We then transfected an increasing dose of either expression plasmid to determine the cooperativity. The results showed that the cyclin D2 promoter activity was dramatically downregulated by EBNA3C and Bcl6 in a dose-independent manner (Fig. 5B). Additionally, when consistent expression of Bcl6 was combined with increasing levels of EBNA3C, we saw a dramatic shutdown of cyclin D2 luciferase activity, suggesting that Bcl6 is a critical partner of EBNA3C in regulating cyclin D2 gene expression (Fig. 5B). Therefore, these results demonstrate that EBNA3C alone has a minimal impact on the transcription activity of cyclin D2 in EBV-positive B cells and may mainly function at the protein level. The results are consistent with those of our study showing that EBNA3C can downregulate Bcl6 repression (29). Therefore, these results demonstrate that EBNA3C alone has a minimal impact on the transcription activity of cyclin D2 in EBV-positive B cells and that its increased levels were due to activities which regulated the cyclin D2 protein.
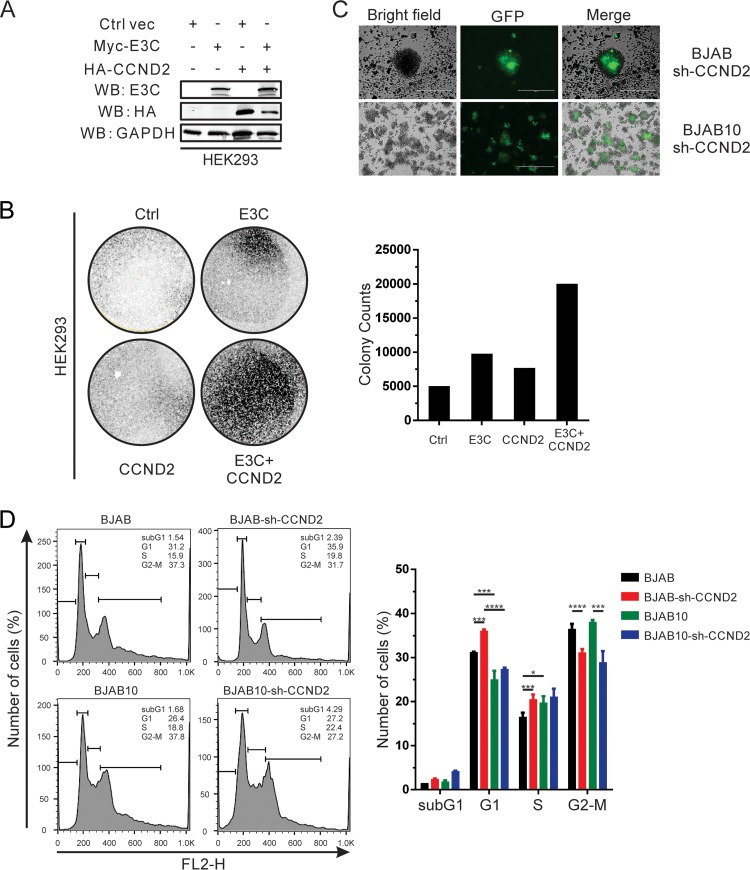
EBNA3C significantly promotes cell proliferation in the presence of cyclin D2.
To further investigate the effects of EBNA3C-mediated cyclin D2 stabilization, HEK293 cells were transfected with EBNA3C, cyclin D2, or both in combination with a green fluorescent protein (GFP) vector (Fig. 6A). These cells were then incubated with G418 for 2 weeks, and the cell colonies were monitored and counted. The results indicate that EBNA3C significantly promotes cell proliferation in cooperation with cyclin D2, suggesting that EBNA3C and cyclin D2 have critical roles in cell proliferation (Fig. 6B). Either molecule had only a minimal impact on the number of colonies seen, but when they were used together, the colony density increased by at least 4-fold, which supports our hypothesis that these two factors cooperate in regulating cell proliferation. To further examine the effects of cyclin D2 in B cells, cyclin D2 knockdown stable cell lines were generated in BJAB and EBNA3C-positive BJAB10 cells (Fig. 6C). Cell cycle analysis was performed with these stable cells. These results showed that cyclin D2 downregulation can inhibit the G1/S transition in BJAB cells but does not affect EBNA3C-associated cell progression in BJAB10 cells stably expressing EBNA3C (Fig. 6D). However, the size of the sub-G1 population in BJAB10 cells in which cyclin D2 was downregulated increased to 2.5-fold compared to that in the isogenic parental BJAB10 cells, suggesting that cyclin D2 can promote cell proliferation by inhibiting cell apoptosis (Fig. 6D). Our results demonstrate that cyclin D2 plays an important role in EBNA3C-mediated cell progression.
FIG 6.
EBNA3C significantly promotes cell proliferation in the presence of cyclin D2. (A) HEK293 cells were cotransfected with Myc-tagged EBNA3C or HA-tagged cyclin D2, and then the selected cells were treated with G418 after 24 h of transfection and selected for 2 weeks. EBNA3C or cyclin D2 expression in these stable cell lines was detected by Western blotting. Ctrl vec, the control vector. (B) After 2 weeks of selection, the numbers of colonies were monitored by quantifying the intensity of the GFP fluorescence. Representative results from two independent experiments are shown. (C) BJAB or BJAB10 cells were infected with lentivirus and treated with puromycin for 3 weeks. Selected cell colonies with GFP fluorescence are shown here. (D) Selected cyclin D2 knockdown B cells were incubated with PI staining buffer and analyzed by flow cytometry. These results were obtained from two independent experiments. sh-CCND2, cyclin D2-specific short hairpin RNA. FL2-H represents the fluorescence of PI. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
DISCUSSION
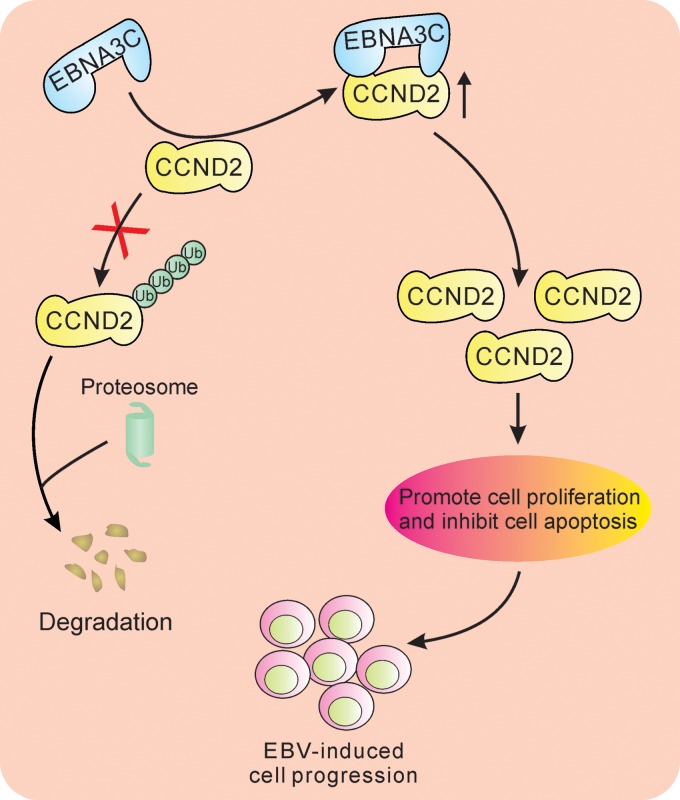
Although EBV has been studied for more than 50 years, the detailed mechanism for EBV-induced lymphomagenesis is still an unsolved mystery. Numerous papers have also focused on the mechanisms of EBV-mediated lymphomagenesis through the control of cell cycle progression. To be specific, latent antigens EBNA2, EBNA-LP, and LMP-1 have been shown to induce cyclin D2 expression during EBV infection (41, 42). This is consistent with our results in EBV-positive cells presented above, in which we detected cyclin D2 mRNA expression in LCL cells (33). In addition, another study using the 4-hydroxytamoxifen (4-HT)-dependent EBNA3C expression model suggested that EBNA3C does not regulate cyclin D2 expression (30). However, several studies have clearly indicated that 4-HT can significantly regulate cyclin D2 expression (43, 44). Therefore, this 4-HT-induced cell model would not fully explain the role of EBNA3C in regulating cyclin D proteins. In this study, we showed that enhanced cyclin D2 upregulation at the protein level is due to EBNA3C expression, which reduced its ubiquitination. EBNA3C also associated with cyclin D, in which they colocalized in the same nuclear compartments, and enhanced its stability by inhibiting the associated ubiquitin degradation pathway. This further adds to the roles of EBNA3C in cell survival and proliferation by regulating another member of the cyclin D family (Fig. 7).
FIG 7.
Schematic model showing the modulation of the cyclin D2 function by EBNA3C. EBNA3C associates with cyclin D2 and enhances its stability at the protein level through inhibition of its ubiquitin-mediated degradation. The EBNA3C-cyclin D2 interaction ultimately drives cell proliferation, which leads to lymphomagenesis.
Cyclin D family members are critical factors involved in regulating G1 progression of the cell cycle. These three cyclin D proteins are highly conserved in human cells, but the coding genes are located at sites on different chromosomes of the human genome (13). Therefore, the complex relationship between these three cyclin D proteins is still unknown because their expression is not always consistent and is strictly controlled. For instance, the expression of cyclins D1 and D2 is different in human B-lymphoid cells (35). Previous studies which checked cyclin D expression in B-cell lymphomas demonstrated that cyclin D2 is predominantly expressed in EBV-transformed lymphoblastoid cell lines (LCLs) and latency III-associated lymphoma cells, while cyclin D3 was expressed in follicular B-cell lymphomas and latency I-associated lymphoma cells (34, 36). The differential expression of cyclin D proteins suggests that their expression is highly regulated and may be switched in EBV-induced lymphomagenesis.
Our study further examined the regulatory network among EBV latent antigens and cyclin D proteins. Previously, cyclin D2 mRNA expression was not enhanced in EBNA3C-positive BJAB cells (33). However, whether EBV can coordinate the expression of the cyclin D proteins in a temporal fashion during cell cycle progression is still to be elucidated. This may involve strict regulatory mechanisms associated with the cell cycle which may control the expression of cyclin D proteins. Interestingly, luciferase reporter experiments showed that the cotransfection of EBNA3C and Bcl6 increased the inhibition of the cyclin D2 promoter activity. However, this effect was not as impressive with increasing amounts of EBNA3C alone. Studies have shown that Bcl6 is a transcription repressor with crucial functions in B-cell development and lymphomagenesis (45). In addition, it can suppress cyclin D2 expression by inhibiting its promoter activity (39, 40, 43, 46). The fact that EBNA3C further enhanced the inhibition of Bcl6-mediated cyclin D2 promoter activity suggests that EBNA3C may also associate with Bcl6 to bind competitively at the promoter or may directly affect Bcl6 expression.
A recent study also showed that viral latent antigens (e.g., EBNA3A, EBNA3C, EBNA2) and other cellular factors (e.g., RBPJ, interferon regulatory factor 4 [IRF4], RUNX3, NF-κB) can contribute to cyclin D2 expression (47). Overall, our work demonstrates that EBNA3C can stabilize the cyclin D2 protein and further promote its ability to mediate cell proliferation. Furthermore, these results indicate the potential value of cyclin D2 as a therapeutic target to treat EBV-induced B-cell lymphomagenesis.
MATERIALS AND METHODS
Ethics statement.
Peripheral blood mononuclear cells (PBMCs) from healthy donors, who provided written and informed consent, were obtained from the Immunology Core of the University of Pennsylvania. The study was also reviewed and approved by the Institutional Review Board (IRB) of the University of Pennsylvania (29).
PBMC infection.
About 10 million cells were incubated with wide-type or EBNA3C-deleted EBV supernatant (multiplicity of infection = 0.1) for 4 h at 37°C. The cells were collected, resuspended in RPMI 1640 medium, and cultured in a 6-well plate. The mRNA levels of the infected cells were determined at the time points postinfection indicated above.
Plasmids, cells, and antibodies.
Myc-tagged and DsRed-tagged EBNA3C and Flag-tagged cyclin D2 have been described previously (33, 48). Hemagglutinin (HA)-tagged cyclin D2 was a gift from Philip Hinds (Addgene plasmid number 8950) (49). The pCDNA3-HA-ubiquitin (Ub) construct was obtained from George Mosialos (Aristotle University of Thessaloniki, Thessaloniki, Greece). HA-tagged Bcl6 was a gift from Riccardo Dalla-Favera (Columbia University, New York, NY). EBNA3C antibody (A10) was a gift from Martin Rowe (University of Birmingham, Birmingham, UK) (50). Myc antibody (9E10) and HA antibody (12CA5) were prepared from hybridomas as previously described (33). Rabbit anti-cyclin D2 polyclonal antibody (C-17) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-Flag antibody (M2) was bought from Sigma-Aldrich (St. Louis, MO). Mouse anti-GAPDH (anti-glyceraldehyde-3-phosphate dehydrogenase) antibody was bought from United States Biological (Swampscott, MA).
HEK293, HEK293T, and Saos-2 cells were kind gifts from Jon Aster (Brigham and Woman's Hospital, Boston, MA). U2OS cells were purchased from the American Type Culture Collection (ATCC). These cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM; HyClone, Logan, UT) supplemented with 5% fetal bovine serum (FBS), 2 mM l-glutamine, 25 U/ml penicillin, and 50 μg/ml streptomycin. EBV-negative BJAB and Ramos Burkitt's lymphoma cells were gifts from Elliot Kieff (Harvard Medical School, Boston, MA). BJAB cells stably expressing EBNA3C (BJAB7 and BJAB10 cells) have been described previously (51). In vitro-transformed EBV-positive LCL1 and LCL2 cells were generated in our laboratory (52). These B-cell lines and PBMCs were cultured in RPMI 1640 medium (HyClone, Logan, UT) and supplemented as described previously (29).
Coimmunoprecipitation (co-IP) and Western blotting (WB).
Transfected HEK293T or Saos-2 cells or the indicated B cells were collected and lysed with radioimmunoprecipitation assay (RIPA) lysis buffer supplemented with protein inhibitors (aprotinin, leupeptin, pepstatin A, and phenylmethylsulfonyl fluoride). Protein quantification was performed using the Bio-Rad protein assay dye reagent concentrate following the manufacturer's instruction. The supernatants were precleared with 30 μl protein A/G Sepharose beads for 1 h at 4°C. Ten percent of the lysate was used as the input, while the rest was incubated with 1 μg specific antibodies and another 30 μl protein A/G Sepharose beads overnight at 4°C. The mixture was then centrifuged and washed three times with ice-cold RIPA buffer. The immune complexes were boiled in Laemmli buffer, fractionated by SDS-PAGE, and transferred onto a 0.45-μm-pore-size nitrocellulose membrane, which was incubated with specific primary and appropriate secondary antibodies. Finally, the signals were captured using an Odyssey infrared imaging system and quantitated with Image Quant software (LiCor Inc., Lincoln, NE).
Immunofluorescence.
Immunofluorescence analysis (IFA) was performed as previously described (29, 53). Briefly, HEK293T, Saos-2, or U2OS cells cultured in 6-well plates were cotransfected with Flag-tagged cyclin D2 and DsRed (red fluorescent protein)-fused EBNA3C. At 48 h posttransfection, these cells were fixed with 4% paraformaldehyde (PFA) and then permeabilized with 0.2% Triton X-100 and blocked with 5% bovine serum albumin in phosphate-buffered saline (PBS). Flag-tagged cyclin D2 was incubated with mouse anti-Flag (M2) antibody followed by Alexa Fluor 488-conjugated secondary antibody (Molecular Probes, Invitrogen, Carlsbad, CA). DsRed-expressing EBNA3C was monitored directly using confocal microscopy. For different B cells, the indicated BJAB, BJAB7, and LCL1 cells were semi-air dried on slides and the immunofluorescence experiments were performed as previously mentioned (29). Endogenous EBNA3C or cyclin D2 was detected with mouse anti-EBNA3C (A10) antibody or rabbit anti-cyclin D2 antibody (Santa Cruz Biotechnology, Santa Cruz, CA), followed by their corresponding secondary antibodies. The nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole; Thermo Fisher Scientific, Waltham, MA). All the slides were visualized on a FluoView FV300 microscope and analyzed with FluoView software (Olympus Inc., Melville, NY).
Real-time PCR.
Total RNA was extracted from the indicated cells using the TRIzol reagent (Invitrogen Inc., Carlsbad, CA) according to the manufacturer's protocol. After treatment with DNase I (Invitrogen Inc., Carlsbad, CA), 2 μg total RNA was reverse transcribed to cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems Inc., Foster City, CA) and then cDNA was mixed with a Power SYBR master mix kit (Applied Biosystems Inc., Foster City, CA). Quantitative real-time PCR was performed, and the results were analyzed using a Step One Plus real-time PCR system (Applied Biosystems Inc., Foster City, CA). Tests with each sample were performed in triplicate, and GAPDH was set as an endogenous control.
Luciferase reporter assays.
HEK293T cells were transfected with the cyclin D2 promoter-driven luciferase reporter and the appropriate combinations of plasmids. The transfected cells were collected after 36 h of transfection, and the luciferase reporter assay was performed using a dual-luciferase reporter system (Promega, USA). A Renilla luciferase-expressing plasmid (pRL-TK) was used as an internal control. All the luciferase activities were measured using an LMaxII384 luminometer (Molecular Devices, Sunnyvale, CA). Representative results from assays performed in duplicate are shown.
Lentivirus production and infection.
Two sense strands of cyclin D2 specifically targeting the coding domain sequence (CDS) of the cyclin D2 gene were designed and cloned into the pGIPZ vector (Addgene, Cambridge, MA) at the XhoI and MluI restriction sites. The corresponding negative-control plasmid including the short hairpin RNA sequence 5′-TCTCGCTTGGGCGAGAGTAAG-3′ (Dharmacon Research, Chicago, IL) was described previously (28). Lentivirus production and infection in order to generate the stable cell lines have been previously described in detail (28).
Stability assay.
About 10 million HEK293T cells were transfected with the constructs indicated above. At 36 h posttransfection, transfected cells were treated with 40 μg/ml cycloheximide (CHX) for the times indicated above. Then, cells were collected, lysed, and analyzed by Western blotting to detect the expression of the indicated proteins.
Colony formation assay.
Approximately 10 million HEK293 cells were transfected with Myc-tagged EBNA3C or HA-tagged cyclin D2 as well as the GFP vector by electroporation. Then, the transfected cells were incubated with G418 at 24 h posttransfection. The GFP fluorescence in a 10-cm culture dish was captured with a PhosphorImager (Molecular Dynamics, Piscataway, NJ) after 2 weeks of selection, and the number of colonies was measured using ImageJ software (NIH, Bethesda, MD). Meanwhile, the selected cells were harvested and the expression of EBNA3C or cyclin D2 was determined by Western blot analysis.
Cell cycle analysis.
Cells were washed with PBS once on ice, fixed in ice-cold 70% ethanol for 30 min at −20°C, washed using PBS, and stained with propidium iodide (PI; 0.5 mg/ml in PBS) plus RNase at 50 μg/ml for 30 min at 37°C. The stained cells were washed again with PBS and analyzed using a FACSCalibur system (Becton Dickinson, Heidelberg, Germany) and FlowJo software (TreeStar, Inc., San Carlos, CA).
Statistical analysis.
The mean values of the data with the standard deviations (SD) are shown in this study. Statistical analyses were performed by Student's two-tailed t test. A P value of <0.05 was considered statistically significant in our results.
ACKNOWLEDGMENTS
We are grateful to George Mosialos (Aristotle University of Thessaloniki, Thessaloniki, Greece), Riccardo Dalla-Favera (Columbia University, New York, NY), Jon Aster (Brigham and Woman's Hospital, Boston, MA), Elliot Kieff (Harvard Medical School, Boston, MA), and Martin Rowe (University of Birmingham, Birmingham, UK) for kindly providing reagents.
This work was supported by the National Cancer Institute at the National Institutes of Health under award numbers P30-CA016520, P01-CA174439, R01-CA171979, and R01-CA177423 (to E.S.R.).
The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Lieberman PM. 2014. Virology. Epstein-Barr virus turns 50. Science 343:1323–1325. doi: 10.1126/science.1252786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young LS, Yap LF, Murray PG. 2016. Epstein-Barr virus: more than 50 years old and still providing surprises. Nat Rev Cancer 16:789–802. doi: 10.1038/nrc.2016.92. [DOI] [PubMed] [Google Scholar]
- 3.Jha H, Pei Y, Robertson E. 2016. Epstein-Barr virus: diseases linked to infection and transformation. Front Microbiol 7:1602. doi: 10.3389/fmicb.2016.01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pei Y, Lewis AE, Robertson ES. 2017. Current progress in EBV-associated B-cell lymphomas. Adv Exp Med Biol 1018:57–74. doi: 10.1007/978-981-10-5765-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Epstein MA, Achong BG, Barr YM. 1964. Virus particles in cultured lymphoblasts from Burkitt's lymphoma. Lancet i:702–703. [DOI] [PubMed] [Google Scholar]
- 6.Middeldorp JM, Brink AA, van den Brule AJ, Meijer CJ. 2003. Pathogenic roles for Epstein-Barr virus (EBV) gene products in EBV-associated proliferative disorders. Crit Rev Oncol Hematol 45:1–36. doi: 10.1016/S1040-8428(02)00078-1. [DOI] [PubMed] [Google Scholar]
- 7.Hammerschmidt W, Sugden B. 1989. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature 340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 8.Cohen JI, Wang F, Mannick J, Kieff E. 1989. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci U S A 86:9558–9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomkinson B, Robertson E, Kieff E. 1993. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol 67:2014–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mannick JB, Cohen JI, Birkenbach M, Marchini A, Kieff E. 1991. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J Virol 65:6826–6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaye KM, Izumi KM, Kieff E. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci U S A 90:9150–9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. 2011. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer 11:558–572. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 14.Harbour JW, Dean DC. 2000. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev 14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 15.Ezhevsky SA, Nagahara H, Vocero-Akbani AM, Gius DR, Wei MC, Dowdy SF. 1997. Hypo-phosphorylation of the retinoblastoma protein (pRb) by cyclin D:Cdk4/6 complexes results in active pRb. Proc Natl Acad Sci U S A 94:10699–10704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ezhevsky SA, Ho A, Becker-Hapak M, Davis PK, Dowdy SF. 2001. Differential regulation of retinoblastoma tumor suppressor protein by G(1) cyclin-dependent kinase complexes in vivo. Mol Cell Biol 21:4773–4784. doi: 10.1128/MCB.21.14.4773-4784.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirzaa GM, Parry DA, Fry AE, Giamanco KA, Schwartzentruber J, Vanstone M, Logan CV, Roberts N, Johnson CA, Singh S, Kholmanskikh SS, Adams C, Hodge RD, Hevner RF, Bonthron DT, Braun KPJ, Faivre L, Riviere J-B, St-Onge J, Gripp KW, Mancini GMS, Pang K, Sweeney E, van Esch H, Verbeek N, Wieczorek D, Steinraths M, Majewski J, FORGE Canada Consortium FC, Boycot KM, Pilz DT, Ross ME, Dobyns WB, Sheridan EG. 2014. De novo CCND2 mutations leading to stabilization of cyclin D2 cause megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome. Nat Genet 46:510–515. doi: 10.1038/ng.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS. 2010. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer 10:59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- 19.Cayrol C, Flemington EK. 1996. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J 15:2748–2759. [PMC free article] [PubMed] [Google Scholar]
- 20.Shore AM, White PC, Hui RC, Essafi A, Lam EW, Rowe M, Brennan P. 2006. Epstein-Barr virus represses the FoxO1 transcription factor through latent membrane protein 1 and latent membrane protein 2A. J Virol 80:11191–11199. doi: 10.1128/JVI.00983-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seto E, Moosmann A, Gromminger S, Walz N, Grundhoff A, Hammerschmidt W. 2010. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog 6:e1001063. doi: 10.1371/journal.ppat.1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinclair AJ, Palmero I, Holder A, Peters G, Farrell PJ. 1995. Expression of cyclin D2 in Epstein-Barr virus-positive Burkitt's lymphoma cell lines is related to methylation status of the gene. J Virol 69:1292–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang S, Willox B, Zhou H, Holthaus AM, Wang A, Shi TT, Maruo S, Kharchenko PV, Johannsen EC, Kieff E, Zhao B. 2014. Epstein-Barr virus nuclear antigen 3C binds to BATF/IRF4 or SPI1/IRF4 composite sites and recruits Sin3A to repress CDKN2A. Proc Natl Acad Sci U S A 111:421–426. doi: 10.1073/pnas.1321704111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian C, Cotter MA II, Robertson ES. 2001. Epstein-Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23-H1: a molecular link to cancer metastasis. Nat Med 7:350–355. doi: 10.1038/85499. [DOI] [PubMed] [Google Scholar]
- 25.Knight JS, Sharma N, Robertson ES. 2005. Epstein-Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc Natl Acad Sci U S A 102:18562–18566. doi: 10.1073/pnas.0503886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saha A, Bamidele A, Murakami M, Robertson ES. 2011. EBNA3C attenuates the function of p53 through interaction with inhibitor of growth family proteins 4 and 5. J Virol 85:2079–2088. doi: 10.1128/JVI.02279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saha A, Lu J, Morizur L, Upadhyay SK, Aj MP, Robertson ES. 2012. E2F1 mediated apoptosis induced by the DNA damage response is blocked by EBV nuclear antigen 3C in lymphoblastoid cells. PLoS Pathog 8:e1002573. doi: 10.1371/journal.ppat.1002573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pei Y, Banerjee S, Sun Z, Jha HC, Saha A, Robertson ES. 2016. EBV nuclear antigen 3C mediates regulation of E2F6 to inhibit E2F1 transcription and promote cell proliferation. PLoS Pathog 12:e1005844. doi: 10.1371/journal.ppat.1005844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pei Y, Banerjee S, Jha HC, Sun Z, Robertson ES. 2017. An essential EBV latent antigen 3C binds Bcl6 for targeted degradation and cell proliferation. PLoS Pathog 13:e1006500. doi: 10.1371/journal.ppat.1006500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maruo S, Wu Y, Ishikawa S, Kanda T, Iwakiri D, Takada K. 2006. Epstein-Barr virus nuclear protein EBNA3C is required for cell cycle progression and growth maintenance of lymphoblastoid cells. Proc Natl Acad Sci U S A 103:19500–19505. doi: 10.1073/pnas.0604919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knight JS, Sharma N, Kalman DE, Robertson ES. 2004. A cyclin-binding motif within the amino-terminal homology domain of EBNA3C binds cyclin A and modulates cyclin A-dependent kinase activity in Epstein-Barr virus-infected cells. J Virol 78:12857–12867. doi: 10.1128/JVI.78.23.12857-12867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knight JS, Robertson ES. 2004. Epstein-Barr virus nuclear antigen 3C regulates cyclin A/p27 complexes and enhances cyclin A-dependent kinase activity. J Virol 78:1981–1991. doi: 10.1128/JVI.78.4.1981-1991.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saha A, Halder S, Upadhyay SK, Lu J, Kumar P, Murakami M, Cai Q, Robertson ES. 2011. Epstein-Barr virus nuclear antigen 3C facilitates G1-S transition by stabilizing and enhancing the function of cyclin D1. PLoS Pathog 7:e1001275. doi: 10.1371/journal.ppat.1001275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pokrovskaja K, Ehlin-Henriksson B, Bartkova J, Bartek J, Scuderi R, Szekely L, Wiman KG, Klein G. 1996. Phenotype-related differences in the expression of D-type cyclins in human B cell-derived lines. Cell Growth Differ 7:1723–1732. [PubMed] [Google Scholar]
- 35.Palmero I, Holder A, Sinclair AJ, Dickson C, Peters G. 1993. Cyclins D1 and D2 are differentially expressed in human B-lymphoid cell lines. Oncogene 8:1049–1054. [PubMed] [Google Scholar]
- 36.Teramoto N, Pokrovskaja K, Szekely L, Polack A, Yoshino T, Akagi T, Klein G. 1999. Expression of cyclin D2 and D3 in lymphoid lesions. Int J Cancer 81:543–550. doi:. [DOI] [PubMed] [Google Scholar]
- 37.Denicourt C, Legault P, McNabb FA, Rassart E. 2008. Human and mouse cyclin D2 splice variants: transforming activity and subcellular localization. Oncogene 27:1253–1262. doi: 10.1038/sj.onc.1210750. [DOI] [PubMed] [Google Scholar]
- 38.Ye W, Blain SW. 2010. S phase entry causes homocysteine-induced death while ataxia telangiectasia and Rad3 related protein functions anti-apoptotically to protect neurons. Brain 133:2295–2312. doi: 10.1093/brain/awq139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. 2000. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity 13:199–212. doi: 10.1016/S1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 40.Dent AL, Vasanwala FH, Toney LM. 2002. Regulation of gene expression by the proto-oncogene BCL-6. Crit Rev Oncol Hematol 41:1–9. doi: 10.1016/S1040-8428(01)00164-0. [DOI] [PubMed] [Google Scholar]
- 41.Arvanitakis L, Yaseen N, Sharma S. 1995. Latent membrane protein-1 induces cyclin D2 expression, pRb hyperphosphorylation, and loss of TGF-beta 1-mediated growth inhibition in EBV-positive B cells. J Immunol 155:1047–1056. [PubMed] [Google Scholar]
- 42.Sinclair AJ, Palmero I, Peters G, Farrell PJ. 1994. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J 13:3321–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandez de Mattos S, Essafi A, Soeiro I, Pietersen AM, Birkenkamp KU, Edwards CS, Martino A, Nelson BH, Francis JM, Jones MC, Brosens JJ, Coffer PJ, Lam EW. 2004. FoxO3a and BCR-ABL regulate cyclin D2 transcription through a STAT5/BCL6-dependent mechanism. Mol Cell Biol 24:10058–10071. doi: 10.1128/MCB.24.22.10058-10071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanda N, Watanabe S. 2004. 17β-Estradiol stimulates the growth of human keratinocytes by inducing cyclin D2 expression. J Invest Dermatol 123:319–328. doi: 10.1111/j.0022-202X.2004.12645.x. [DOI] [PubMed] [Google Scholar]
- 45.Basso K, Dalla-Favera R. 2012. Roles of BCL6 in normal and transformed germinal center B cells. Immunol Rev 247:172–183. doi: 10.1111/j.1600-065X.2012.01112.x. [DOI] [PubMed] [Google Scholar]
- 46.Hirata Y, Ogasawara N, Sasaki M, Mizushima T, Shimura T, Mizoshita T, Mori Y, Kubota E, Wada T, Tanida S, Kataoka H, Kamiya T, Higashiyama S, Joh T. 2009. BCL6 degradation caused by the interaction with the C-terminus of pro-HB-EGF induces cyclin D2 expression in gastric cancers. Br J Cancer 100:1320–1329. doi: 10.1038/sj.bjc.6605010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt SC, Jiang S, Zhou H, Willox B, Holthaus AM, Kharchenko PV, Johannsen EC, Kieff E, Zhao B. 2015. Epstein-Barr virus nuclear antigen 3A partially coincides with EBNA3C genome-wide and is tethered to DNA through BATF complexes. Proc Natl Acad Sci U S A 112:554–559. doi: 10.1073/pnas.1422580112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jha HC, Lu J, Saha A, Cai Q, Banerjee S, Prasad MA, Robertson ES. 2013. EBNA3C-mediated regulation of Aurora kinase B contributes to Epstein-Barr virus-induced B-cell proliferation through modulation of the activities of the retinoblastoma protein and apoptotic caspases. J Virol 87:12121–12138. doi: 10.1128/JVI.02379-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baker GL, Landis MW, Hinds PW. 2005. Multiple functions of D-type cyclins can antagonize pRb-mediated suppression of proliferation. Cell Cycle 4:330–338. doi: 10.4161/cc.4.2.1485. [DOI] [PubMed] [Google Scholar]
- 50.Maunders MJ, Petti L, Rowe M. 1994. Precipitation of the Epstein-Barr virus protein EBNA 2 by an EBNA 3c-specific monoclonal antibody. J Gen Virol 75(Pt 4):769–778. doi: 10.1099/0022-1317-75-4-769. [DOI] [PubMed] [Google Scholar]
- 51.Robertson ES, Grossman S, Johannsen E, Miller C, Lin J, Tomkinson B, Kieff E. 1995. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein J kappa. J Virol 69:3108–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cotter MA II, Robertson ES. 2000. Modulation of histone acetyltransferase activity through interaction of Epstein-Barr nuclear antigen 3C with prothymosin alpha. Mol Cell Biol 20:5722–5735. doi: 10.1128/MCB.20.15.5722-5735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pei Y, Fu W, Yang E, Shen A, Chen YC, Gong H, Chen J, Huang J, Xiao G, Liu F. 2012. A Hsp40 chaperone protein interacts with and modulates the cellular distribution of the primase protein of human cytomegalovirus. PLoS Pathog 8:e1002968. doi: 10.1371/journal.ppat.1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]